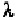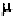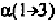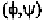Biology Reference
In-Depth Information
167
With these restrictions and other information derived from mutational
studies, symmetry of tetramer subunits, etc., the three-dimensional structure
of the backbone of ribitol dehydrogenase (Stevens and Wu, 1985) has been
proposed based on the predictive method discussed in this Chapter. While it
is not sufficient to select angles for each amino acid residue of a
protein to predict its three-dimensional folding, the incorporation of
additional information can, in some cases, provide sufficient restrictions to
predict the tertiary structure of the backbone of a protein.
With the rapid development of various genome projects, the amino acid
sequences of many proteins, some represented by the so-called open reading
frames (ORF's), will become available. Prediction of their three-
dimensional folding and possible structures can provide valuable biological
information to their functions.
SIDE-CHAIN STRUCTURE PREDICTION
After the backbone structure of a peptide loop, e.g. one of the CDR's of
antibodies, is proposed, the side chains of all amino acid residues in the loop
can be analyzed. An example is illustrated for an immunoglobulin,
MOPC104E (McIntire
et al.,
1965). It is an IgM with a light chain
Appella
et al.,
1968) and a heavy chain (Kehry
et al.,
1979). It reacts with
dextran, especially dextran B1355 from
L. mesenteroides,
thus specific for
the linkage. Nigertriose can very effectively inhibit MOPC104E's
binding of dextran B1355, suggesting that the binding site may consist of
three residues of -linked glucose. Further study (Schepers
et al.,
1978) suggests that there is a 12 A-deep cavity complementary to the
nigerose disaccharide with a subsite for a third glucose residue in an
adjacent groove. MOPC104E is thus specific for terminal nigerosyl residues
as in the case of dextran B1355, but not for
linkages only in the
middle of dextran molecules.
Various attempts to crystallize MOPC104E for X-ray diffraction studies
have unfortunately failed. Thus, its detailed antibody combining site three-
dimensional structure can only be predicted (Hovis and Wu, 1985). Similar
to MOPC315, the predicted CDR's are joined onto the framework structure
of Newm, except in the region of CDRL2 where that of REI (Epp
et al.,
1975) is also used. In the order of prediction, the CDR amino acid
sequences and their
angles are listed in Tables 6-7 to 6-12 (Hovis,
1982).