Biology Reference
In-Depth Information
signature
algorithm
BLAST
organism A
organism B
organism B
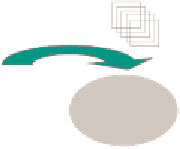
Fig. 4.
Starting from a set of coexpressed genes (yellow dots in left box) associated
with a particular function in organism A, we first identify the homologes in organism
B using BLAST (middle box). Only some of these homologs are coexpressed, while
others are not (blue dots). The signature algorithm selects this coexpressed subset,
and adds further genes (light yellow) that were not identified based on sequence but
share similar expression profiles (right box).
to seeds containing the orthologs of coexpressed yeast genes with known
cellular functions (Fig. 4). In most cases, a coexpressed subset of these
orthologs could be identified. These genes are likely to participate in a
function similar to that of the original yeast genes. Moreover, this approach
provides functional predictions for genes that have similar expression pat-
terns but no sequence similarity with the original genes.
The modular structures of the expression data were characterized
by first identifying the transcription modules in each dataset (using the
ISA) and subsequently their organization in each transcription pro-
gram (c.f. Fig. 8). The coexpression of several sets of ancient genes
pertaining to fundamental cellular functions (like those coding for
ribosomal proteins) has been conserved across all organisms. Yet,
the relative importance of these conserved modules to the transcrip-
tion program varies significantly between organisms. Moreover, a sig-
nificant number of modules are composed primarily of genes that are
organism-specific.
35
Comparing the coexpression of modules (rather than of individual
genes), we revealed similarities and differences in higher-order structures of
the various transcription programs. Specifically, we asked whether pairs of
modules that are (anti)correlated in one organism exhibit the same regula-
tory relationship in another organism. Studying a set of eight representa-
tive modules related to core cellular processes among the six diverse