Biology Reference
In-Depth Information
sequences with experimental
motif annotations
performance
indices
superposition of
experimental and
predicted motifs
motif description
(consensus sequence
hide annotations
sequences with predicted
or weight matrix)
motif annotations
motif
discovery
program
motif
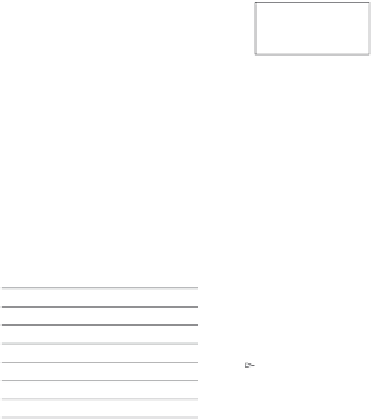
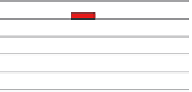
Fig. 3.
Benchmarking protocol for evaluation of motif discovery algorithms. A test
consists of DNA sequences which experimentally mapped binding sites to a particu-
lar transcription factor (experimental motif annotations are shown in red). The naked
sequences (without motif annotations) are given as input to the motif discovery algo-
rithm, which returns a set of predicted motif instances plus, optionally, a motif
description. The experimental and predicted motif annotations are then superim-
posed for computation of a variety of performance indices, such as sensitivity and
specificity. Partial overlaps between known and predicted motifs are usually counted
as success. This protocol has been applied in the recent benchmarking studies
described in Hu
et al
.
29
and Tompa
et al
.
30
annotations, of course, are hidden to the program. The task of the
motif discovery program is to rediscover the hidden motif and to
return the coordinates of the corresponding motif instances. The
performance is evaluated on the basis of the overlap between experi-
mental and predicted motif annotations, and is expressed by standard
measures such as sensitivity (percentage of true motif instances over-
lapped by predicted motif instances) and specificity (percentage
of predicted motif instances overlapped by true motif instances).