Biology Reference
In-Depth Information
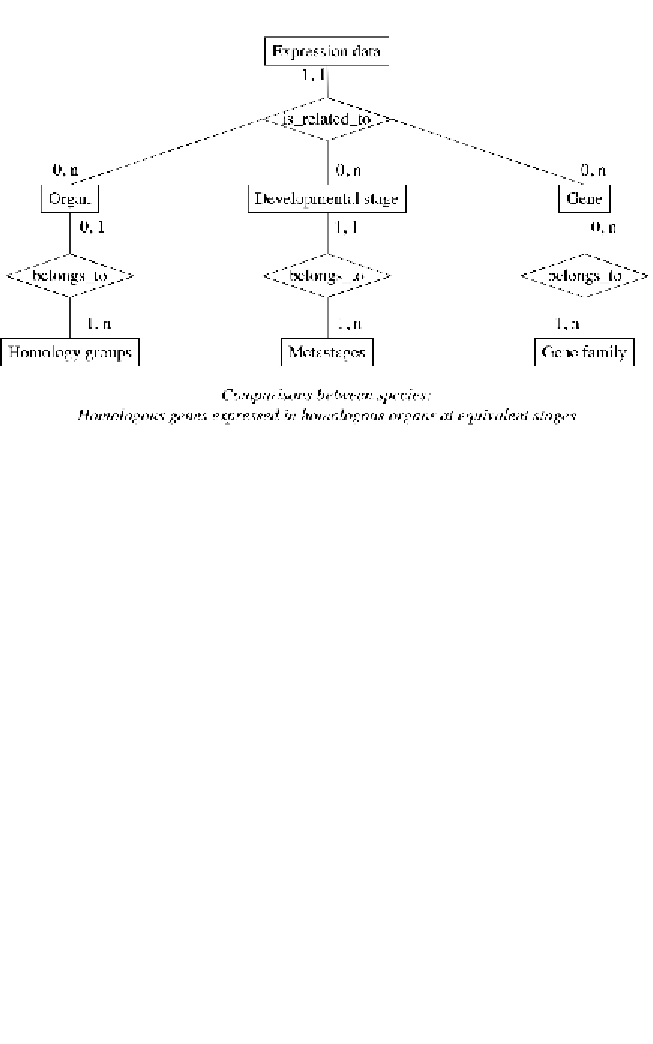
Fig. 4.
Schematic representation of the relationships between types of information
in the Bgee database. Expression data are central to relating genes to anatomical and
developmental terms in each species.
We therefore developed a simplified ontology of metastages. Despite the
resulting loss of accuracy, it allows comparison of gene expression pat-
terns, taking into account developmental time.
Concerning expression data, we face two challenges: integrating het-
erogeneous data types,
85,86
and transforming quantitative data (level of
expression) into the qualitative information that is standard in typical
developmental studies (“expressed” or not). The reason for integrating
heterogeneous expression data is that they complement each other in
terms of coverage. For example, EST libraries typically present an incom-
plete picture of the transcriptome, but they are available for many species
and allow good identification of closely related paralogs. Oligonucleotide
microarrays are much more complete, but different experiments are dif-
ficult to compare and nonmodel species are not covered. Both ESTs and
microarrays are usually annotated to coarse anatomical and developmen-
tal descriptions (e.g. “adult brain”), whereas
in situ
hybridizations can
provide very detailed accounts of gene expression. However,
in situ
hybridization provides limited genome coverage (although see Thisse