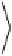Biology Reference
In-Depth Information
This leads to
=
Ú
dl
H
()
l
(14)
B
,
D
G
d
l
AB
dl
l
A
l
which is the TI formula.
22,30
Note that the FEP formula [Eq. (13)] can
be recovered from Eq. (14) by considering a first-order numerical
approximation of the Hamiltonian derivative.
31
In practice, simulations
are performed at a number of fixed
λ
values between and including
λ
A
and
) is calculated and
the phase space average in Eq. (14) is estimated. In the end, the integra-
tion over
λ
B
, during which the analytical derivative of
H
(
λ
is performed numerically. Note that the same care has to be
taken as for FEP in choosing the
λ
λ
dependence of
H
(
λ
) in order to avoid
numerical singularities and optimize convergence.
The recent adaptive integration method
32
seeks to estimate the same
integral as TI [Eq. (14)]. In addition to fixed
sampling, it uses a
Metropolis Monte Carlo procedure to generate moves that change the
value of
λ
during the simulation. This method seems to be one of the
most efficient to date.
33
λ
4.2. Relative Free Energy Differences from
Thermodynamic Cycles
A common application of MD free energy calculations is to compute the
relative binding free energy of two ligands
L
1
and
L
2
to a receptor
R
. In
this case, one can avoid the computationally difficult task of computing
directly the binding free energy of each ligand,
∆
G
1
and
∆
G
2
, by using
the thermodynamic cycle depicted in Fig. 5.
Since free energy is a state function, the difference of the horizontal
legs is equal to the difference of the vertical legs in Fig. 5:
DD
GGGG G
=
D
-
D
=
D
-
D
solv
.
12
2
1
bind
Therefore,
∆∆
G
12
can be obtained by calculating the solvation free energy
difference
∆
G
solv
and the receptor interaction free energy difference