Biology Reference
In-Depth Information
hormone treatment
Both the ga1-3 gai-t6 rga-t2 rgl1-1 (Q3) and opr3 mutant plants (~27 days old)
were sprayed with mock (0.1% ethanol v/v), GA3 (10?4 M) (Sigma) or MeJA
(0.015% v/v) (Sigma). After treatment, young inflorescences were collected at dif-
ferent time point (18 hrs, 48 hrs, 72 hrs and 96 hrs) for total RNA extraction. For
observing rescue of stamen development, mutant plants were repeatedly treated
(once a week) with GA or JA.
RT-PCR and Northern Analysis
Different organs (sepal, petal, stamen, pistil and peduncle) of stage 11-12 flow-
ers were dissected under microscope and pooled for RNA extraction. Flowers
younger than stage 11 were pooled as young flower buds for RNA extraction. To-
tal RNA was extracted from the young flower buds of respective genotypes treated
with or without GA and JA using Tri Reagent (Molecular Research Center, Cin-
cinnati, OH). The residue genomic DNA in the total RNA was removed via
treatment with RNase-free DNase I (Roche, Germany) and the total RNA further
purified with the RNeasy Mini kit (QIAgen, Valencia, CA, USA). First strand
cDNA was synthesized using SuperScript™II RNase H? Reverse Transcriptase
(Invitrogen, USA). First strand cDNA was used as the template in PCR using
gene specific primers. Primer pairs used in identification of DELLA-repressed
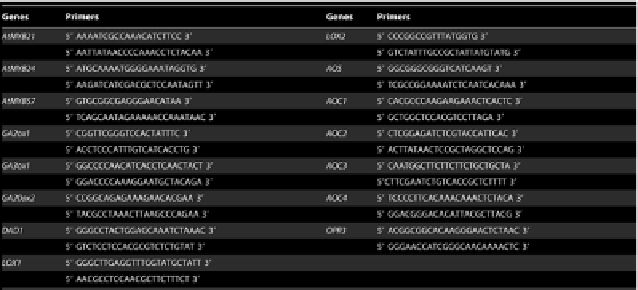
stamen-enriched genes were listed in Table S1. Primer pairs for RT-PCR analysis
of GA2ox1, GA3ox1, GA20ox2, DAD1, AOS, OPR3, LOX1, 2 and AOC1, 2,
3, 4 were listed in Table 4. For quantifying the gene expression levels, PCR prod-
ucts were stained with ethidium bromide and the intensity was quantified using
software Molecular Analyst (Bio-Rad). The gene expression level was normalized
to the expression level of ACTII and then displayed as a ratio of expression levels
of GA (or JA) treated samples versus untreated control.
Table 4.
Gene specific primer pairs used in RT-PCR analysis.