Information Technology Reference
In-Depth Information
jETI helper SIBs
provide basic functionality for loading data from differ-
ent sources so that it can be transferred to the jETI server(s), as well as
for retrieving and further processing the returned results.
The workflow developer can deliberately make use of the framework, the plu-
gins and the readily provided SIBs as described above. In addition, concrete
jABC workflow projects comprise:
•
1.
application-specific SIB libraries
, providing the functionality that is re-
quired for the developed workflows, but not provided by the standard set
of SIBs,
2. PROPHETS
domain models
as described in detail in Section 2.3.2, com-
prising service and type taxonomies, semantic service annotations and
constraints, and
3. the actual
SLGs
that implement the workflows.
Focusing on the application of the framework in the bioinformatics ap-
plication domain, the development of workflow applications is in fact what
constituted the main part of the work described in this topic. This becomes
evident in particular in Part II, where the selected bioinformatics application
scenarios are described in detail, comprising the provisioning of specific SIB
libraries, the definition of semantic domain models, and the actual workflow
design.
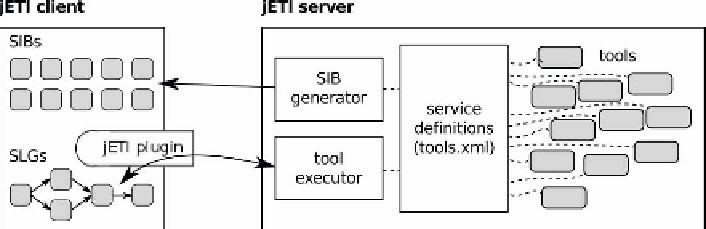
2.1.2 The jETI Platform
The jETI platform [301, 202, 203] is an easy-to-use tool integration technology
that is tailored to making file-based applications remotely accessible. It has
been name-giver for Bio-jETI, since the integration of all kinds of existing
analysis tools in order to provide them in a form that is easily accessible
by workflow frameworks (like the jABC) is central to the rigorously service-
oriented approach to bioinformatics workflow management that has been
followed by this work.
Fig. 2.5
Schematic overview of the jETI framework



Search WWH ::

Custom Search