Chemistry Reference
In-Depth Information
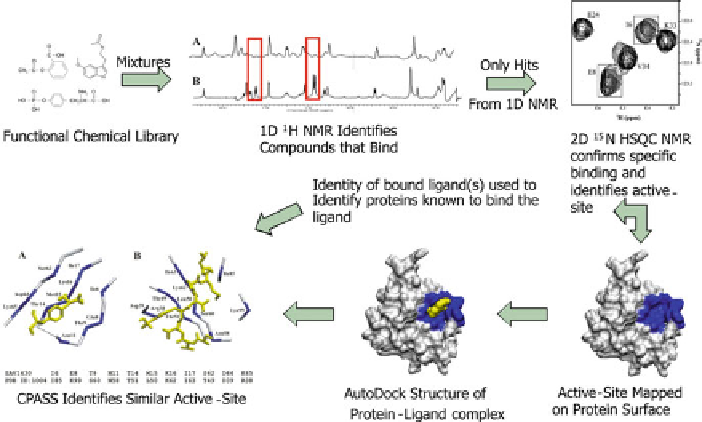
Fig. 7 A flow diagram of the FAST-NMR process. Mixtures of biologically active compounds are
first assayed in a ligand-based 1D line broadening screen against the protein of interest.
Compounds that are identified as hits are then verified using CSPs from a 2D
1
H-
15
N HSQC
experiment that define a binding site on the protein surface. The CSPs are used to guide and filter
an AutoDock molecular docking calculation to generate a protein-ligand co-structure. The ligand
binding site defined by the co-structure is then compared to other experimental binding sites in the
PDB using CPASS. (Reprinted with permission from [
28
], copyright 2008 by Elsevier)
potential binders. These hits are then verified in a target-based screen using a 2D
1
H-
15
N HSQC experiment, where the occurrence of CSPs allows for the identifica-
tion of the ligand binding site. Molecular docking is used to generate a rapid
protein-ligand co-structure [
121
] that serves as input for the Comparison of Protein
Active-Site Structures (CPASS) program [
153
]. CPASS compares the sequence
and structure of this NMR modeled ligand binding site to ~36,000 unique experi-
mental ligand binding sites from the RCSB Protein Databank [
143
]. Thus, a protein
of unknown function can be annotated from a protein with a known function that
shares a similar ligand binding site [
154
]. The FAST-NMR and CPASS approach
has been used for the successful annotation of two hypothetical proteins, SAV1430
from
S. aureus
[
29
] and PA1324 from
P. aeruginosa
[
155
]. It has also been used
to identify a structural and functional similarity between the bacterial type III
secretion system and eukaryotic apoptosis [
156
].
The FAST-NMR approach was recently applied to protein YndB from
Bacillus
subtilis
to generate a functional annotation [
112
]
.
FAST-NMR was augmented
by the inclusion of a virtual screen using the Nature Lipidomics Gateway library
that contains ~22,000 lipids. Eight major categories of lipids are represented
in the library (fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterol
lipids, prenol lipids, saccharolipids, and polyketides), which are further divided into
a total of 538 distinct subclasses. The initial goal was to identify lipid scaffolds that