Chemistry Reference
In-Depth Information
Dehydration, followed by addition of the organic phase, allows micelles to “flip” out
into a reverse micelle configuration while maintaining the hydrophobic domain of the
membrane protein in a membrane-mimetic environment. In this complex, each mem-
brane protein is thought to be associated with two reverse micelles that converge
around the hydrophobic domain of the protein (Fig.
3
)[
225
,
227
,
229
]. In some cases
co-surfactants (e.g., dihexadecyldimethylammonium bromide (DHAB)) and/or co-
solvents (e.g., hexanol) are also required to stabilize this structure. This approach was
shown to be successful using the tetrameric KcsA channel as a test system [
228
].
Reconstitution in reverse micelles was achieved by purification in CTAB, followed by
lyophilization and addition of DHAB, hexanol, pentane, and water. The result was a
well-resolved
1
H-
15
N HSQC spectrum for KscA, with significantly enhanced trans-
verse relaxation times relative to those in water (80 ms vs 20 ms). Moreover,
potassium ion-dependent chemical shifts in the channel selectivity filter could be
observed (Fig.
4
), confirming a functional state for this sample. Although protein
concentrations in reverse micelles tend to be lower than what can be obtained in
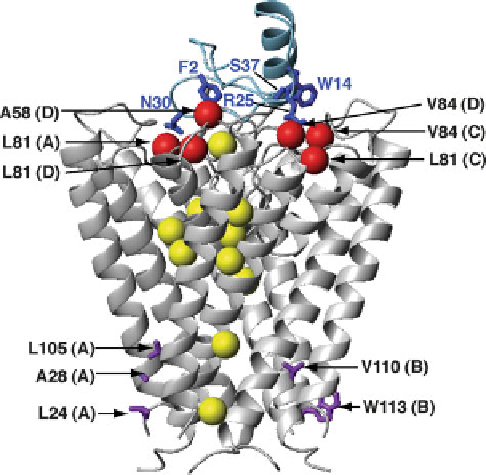
Fig. 4 Novel insights into KcsA potassium channel structure and function revealed by solution
NMR. A ribbon diagram representation of the complex determined for the KcsA tetramer (
gray
)
bound to charybdotoxin (
blue
)[
298
]. Highlighted are KcsA methyl groups (
red balls
) that gave
rise to intermolecular NOEs with charybdotoxin side chains (
sticks
), showing the importance of
methyl groups in defining this complex. Selectivity filter residues that were observed to give rise to
potassium-dependent chemical shifts are shown in
purple
[
228
,
273
]. In addition, methyl groups of
side chains showing the largest chemical shift differences between active and resting states are
shown in
yellow
for one subunit (residues L24, L40, L59, V70, V76, V91, V95, I100, and L105),
indicating widespread changes throughout the TM region between the two states [
273
]. Residue
labels include KcsA subunits A, B, C, and D in parentheses. All ribbon structure figures were made
with MOLMOL [
398
]