Biology Reference
In-Depth Information
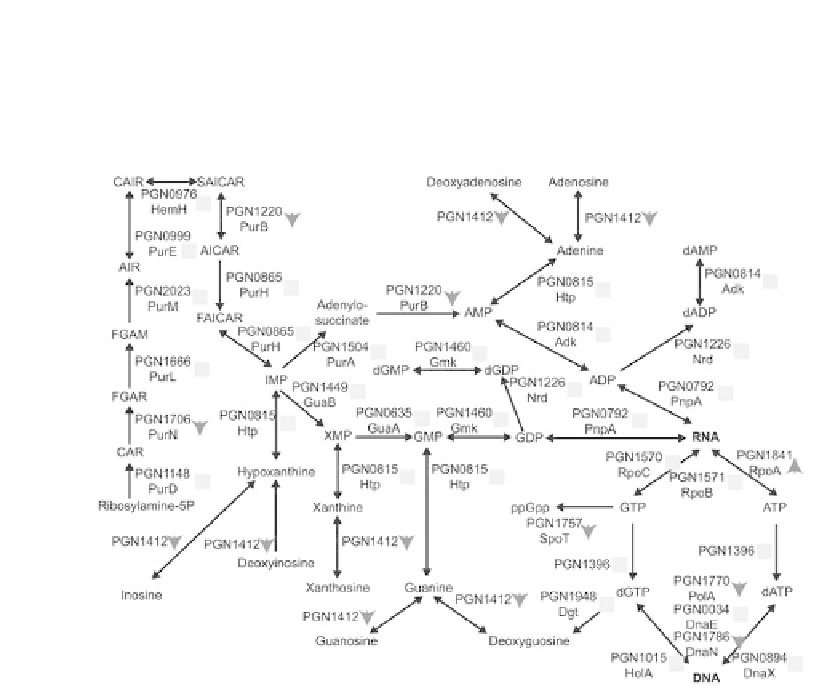
Purine biosynthesis does not appear to be signifi cantly effected in the three spe-
cies community (Figure 6). A few proteins showed reduced abundance, but the central
biosynthesis pathway was primarily unchanged.
Figure 6.
Purine biosynthetic pathway, showing protein abundance changes for the
P. gingivalis-F.
nucleatum-S. gordonii
/
P. gingivalis
comparison. The protein names and arrows/squares follow the
same conventions as in Figure 5. The RNA and DNA are shown in bold. GAR: 5-Phosphoribosyl
glycinamide; FGAM: 5-phosphoribosyl-N-formylglycineamidine; FGAR: 1-(5'-Phosphoribosyl)-N-
formylglycinamide; AICAR: 5'-phosphoribosyl-4-(N-succinocarboxamide)-5-aminoimidazole; AIR:
1-(5'-Phophoribosyl)-5-aminoimidazole; CAIR: 5'P-Ribosyl-4-carboxy-5-aminoimidazole; SAICAR:
5'P-Ribosyl-4-(N-succinocarboximide)-5-aminoimidazole; FAICAR: 1 (5'-Phosphoribosyl)-5-
formamido-4-imidazole carboxamide.
Stress Proteins
The ability of the community to provide physiologic support to constituent species
might result in
P. gingivalis
experiencing lower levels of environmental stress than
occurs in monoculture. Consistent with this concept, community derived
P. gingiva-
lis
showed a significant reduction in abundance of DNA repair proteins (PGN0333,
RadA; PGN0342, Ung; PGN0367, Xth; PGN1168, MutS; PGN1316, UvrA;
PGN1388, LigA; PGN1567, RecF; PGN1585, UvrB; PGN1712, Nth; PGN1714,
Mfd; PGN1771, Pol1). The DNA repair genes are generally induced in the presence
of damaged DNA [41], and lower abundance of DNA repair proteins is consistent
with the monoculture experiencing more DNA damage than
P. gingivalis
in the three
species community where the presence of the partner organisms provides protection
against DNA damage.