Biology Reference
In-Depth Information
In this respect, here we report on a new cold-adapted β-D-galactosidase, isolated
from psychrothrophic, Antarctic
Arthrobacter
sp. 32c bacterium strain, that possesses
low molecular weight of 75.9 kDa of monomer and 195 kDa of native protein. In addi-
tion, the presented enzyme is active in the range of temperature 4-8°C that is suitable
for milk industry applications and can be produced extracellularly on a large scale us-
ing recombinant
P. pastoris
strains cultivated either on methanol or glycerol (a cheap
by-product in biodiesel industry).
Characterization of 32c Isolate
Many different colonies were isolated from the Antarctic soil. One isolate, named
32c, that formed yellow colonies was chosen for further study because of its ability
to hydrolyze X-Galthe cromogenic analogue of lactose. The cells were gram-nega-
tive rods. The optimum growth in LAS medium was observed between 25-27°C. No
growth occurred at 37°C. In order to determine the ability of the selected isolate to
utilize starch, milk, avicell, or arabinose several plates with different substrates were
prepared. It was observed that 32c strain produces enzymes of industrial interest like
α-amylase, proteases and has an arabinose utilization pathway. In order to estimate
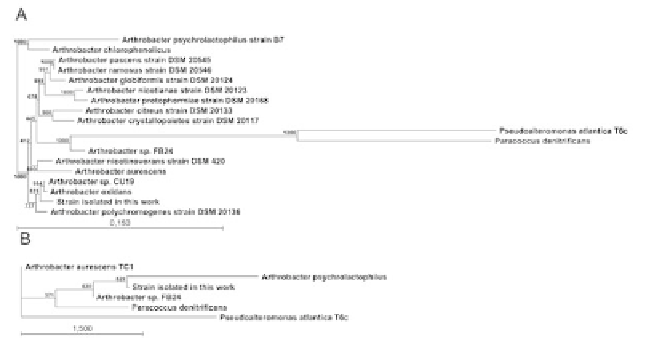
the phylogenetic position of the isolate, we cloned the amplified 16S rRNA gene into
pCR-Blunt vector, determined its sequence, and examined its phylogenetic relation-
ships (Figure 1A). The obtained sequence was deposited at GenBank with the acces-
sion no. FJ609656. An analysis of the sequence showed that it clustered with other or-
ganisms isolated from cold environments, mainly belonging to
Arthrobacter
species.
The isolate formed a well-defined cluster with
A. oxidans
(98.59% sequence identity)
and
A. polychromogenes
(97.86% sequence identity). Based on 16S rDNA similarity,
physiological properties similar to other
Arthrobacter
strains and its presence in the
Antarctic soil our isolate was classified as
Arthrobacter
sp. 32c.
Figure 1.
Phylogenetic analysis of the
Arthrobacter
sp. 32c 16S rDNA sequence (A) and
Arthrobacter
sp. 32c
-D-galactosidase gene sequence (B). Sequences were aligned using the sequence analysis
softwares: ClustalX 1.5 b and Gene-Doc 2.1.000. Phylogenetic trees were reconstructed with
the PHYLIP COMPUTER PROGRAM PACKAGE, using the neighbor-joining method with genetic
distances computed by using Kimura's 2-parameter mode. The scale bar indicates a genetic distance.
The number shown next to each node indicates the percentage bootstrap value of 100 replicates.
β