Biology Reference
In-Depth Information
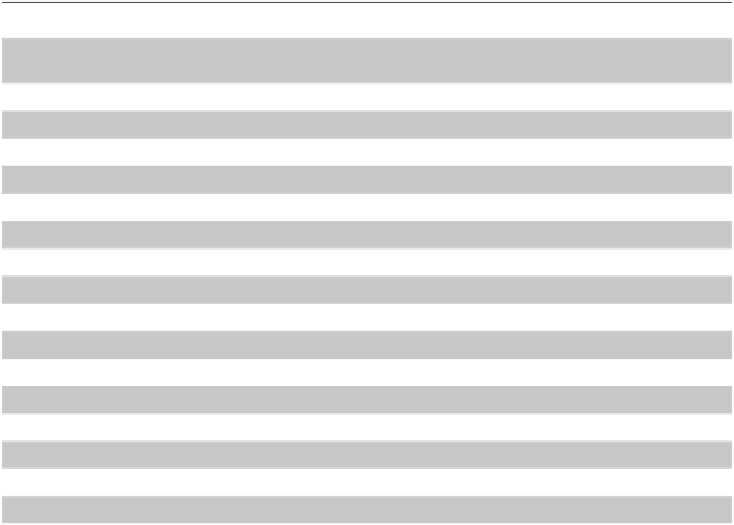
Table 1.
Summary of the main characteristics of the iJP815 metabolic model.
System
Parameter
Subset
Size
P. putida
KT2440
Genome size
6.18 Mbp
Total ORFs
5446
iJP815
Reactions
Total
877
Potentially active
588 (67.0%)
Unconditionally Blocked
289 (33.0%)
Well annotated
764 (87.1%)
Weakly annotated
57 (6.5%)
Non-gene-associated
56 (6.4%)
Transport
70 (8.0%)
Metabolites
Total
888
Internal
824 (92.8%)
Balanced
461 (55.9%)
Unbalanced
363 (44.1%)
External
64 (7.2%)
Genes
Total
815
Well annotated
701 (86.0%)
Weakly annotated
114 (14.0%)
The fi nal reconstruction accounts for the function of 815 genes, corresponding to
15% of all genes in the
P. putida
genome and to 65% (1,253) of those currently as-
signed to the classes “Metabolism” (K01100) and “Membrane Transport” (K01310)
in the Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology classifi cation
[35]. These fi gures are consistent with recently published metabolic reconstructions
for other prokaryotes.
Model Assessment and Extension Through High-throughput Phenotyping
Assays
A high-throughput BIOLOG phenotypic assay was performed on
P. putida
to validate
and extend the model. In this assay,
P. putida
was tested for its ability to oxidize 95
carbon substrates in minimal medium. Of these 95 substrates,
P. putida
oxidized 45.
We added two other carbon sources to the positive-oxidation group (L-phenylalanine
and L-threonine) despite a negative BIOLOG result, since these substrates had been
previously shown to be growth substrates [16] and since we confirmed these results
experimentally (data not shown), giving altogether 47 compounds utilized
in vivo
.
Forty-seven out of the 95 carbon sources tested were accounted for in iJP815
pre2
, en-
abling a comparison of these BIOLOG data with FBA simulations of iJP815 grown
on
in silico
minimal medium with the respective compound as sole carbon source (see
Table 2).