Biology Reference
In-Depth Information
and (3) a model completion stage in which BIOLOG substrate utilization data was
used to guide model expansion and
in silico
viability on varied substrates. The final
reconstruction, named iJP815 following an often used convention [33], consists of 824
intracellular and 62 extracellular metabolites connected by 877 reactions. Eight hun-
dred twenty-one (94%) reactions have at least one assigned gene as delineated in the
gene-protein-reaction (GPR) relationships. The GPR relationships are composed of
Boolean logic statements that link genes to protein complexes and protein complexes
to reactions via combinations of AND and OR operators. An “AND” operator denotes
the required presence of two or more genes for a protein to function (as in the case of
multi-protein complexes), while an “OR” operator denotes a redundant function that
can be catalyzed by any of several genes (as in the case of isozymes). Only 56 reac-
tions, of which nine are non-enzymatic, lack associated genes. The remaining 47 non-
gene-associated, enzymatic reactions were added in order to close metabolic network
gaps identified during the successive steps of the reconstruction process.
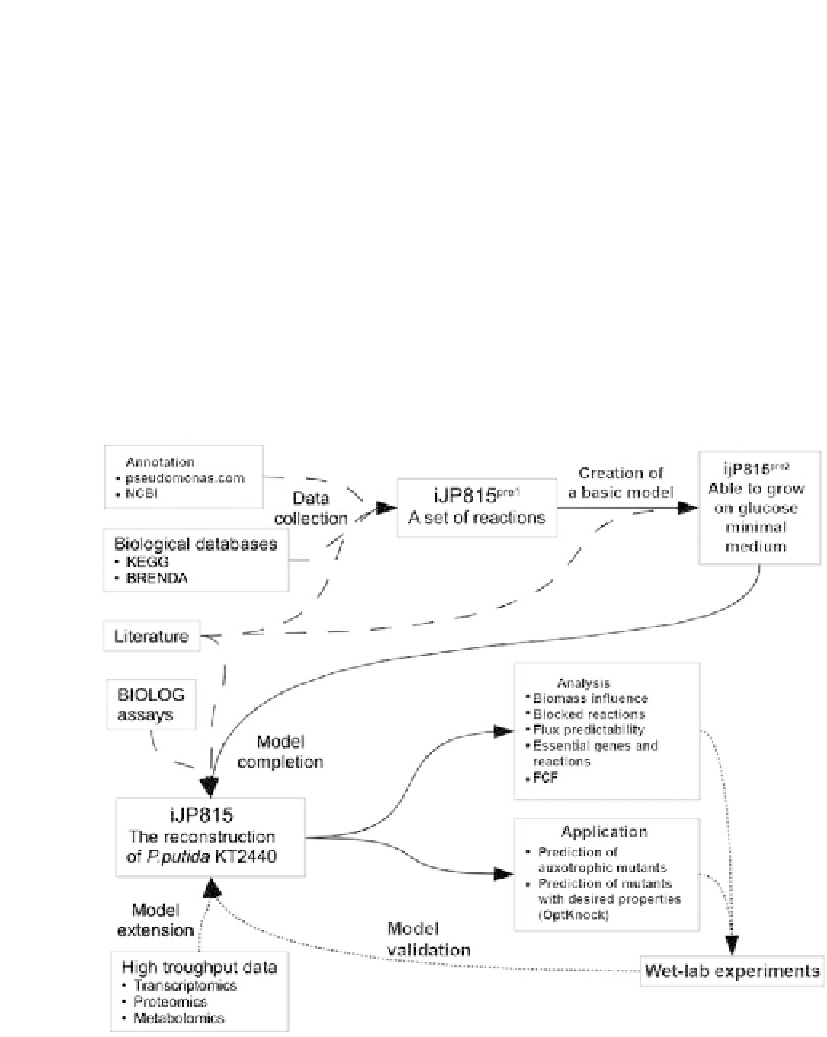
Figure 1.
Schematic diagram of the metabolic reconstruction and analysis processes.
Solid lines
indicate consecutive steps of the reconstruction. Dashed lines represent information transfer. Dotted
lines specify planned tasks.
Most network gaps (27) were identifi ed during the second round of the reconstruc-
tion and were resolved through detailed literature mining, thereby enabling iJP815 to