Biology Reference
In-Depth Information
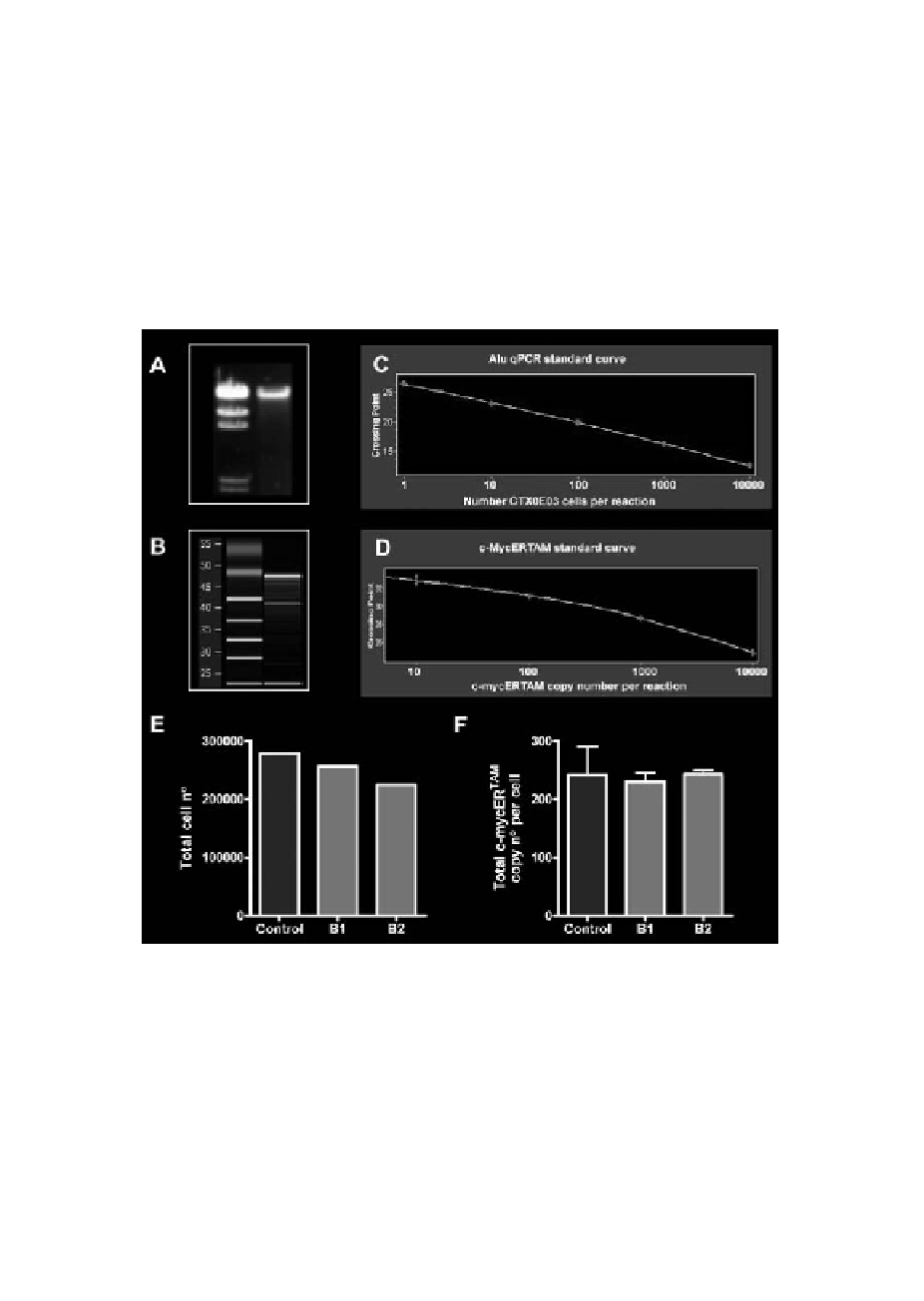
prior to delivery contained 46,600 cells/
µ
l - i.e. total of 279,600 cells were pres-
ent in 6
µ
l loaded into the syringe. By Alu sequence analysis, the injected rat brain
tissue sections contained an average of 241,116 (SEM = 15,487) CTX0E03 cells
(Figure 1F). The measured number of CTX0E03 cells is within the range expect-
ed. Analysis of the total RNA isolated from these samples showed CTX0E03 cells
to express on average 236.9 (SEM = 7.8) c-mycERTAM transcript copies per cell
(Figure 1G). Control CTX0E03 cells measured in culture were shown to have a
similar value of 242.3 (SEM = 47.8) c-mycERTAM transcript copies per cell.
Figure 1.
Alu sequence and c-mycER
TAM
assay development and validation. Gels showing the quality and purity
of gDNA by agarose gel electrophoresis (A) and RNA by virtual gel produced by Agilent 2100 Bioanalyzer (B,
RNA Integrity Number >9.4 as analyzed by Agilent RNA 600 nano kit [32]) isolated from the same sample.
Standard curves used to determine: cell number by Alu sequence qPCR (C, Error 0.0300, efficiency 1.993;
3 replicates); absolute c-mycER
TAM
copy number by c-mycER
TAM
qRT-PCR (D, Error 0.0837 and efficiency
2.131; 3 replicates). All standard curves were generated from CTX0E03 gDNA diluted in rat gDNA or cDNA.
Crossing point refers to the number of PCR cycles required to generate a detectable fluorescent signal generated
on a Roche LC480 instrument. Positive control rat brain samples (B1 and B2) were grafted with approximately
300,000 CTX0E03 cells each and harvested immediately (E, F). Data shown is the total number of CTX0E03
cells in each tissue section as determined by Alu, where control is the number of viable cells in the cell suspension
prior to injection as determined by counting using a haemocytometer (E); and total c-mycER
TAM
transcript copy
number calculated per CTX0E03 cell detected in brain samples, where control is the number of copies per cell
detected in vitro culture (F).
Search WWH ::

Custom Search