Biology Reference
In-Depth Information
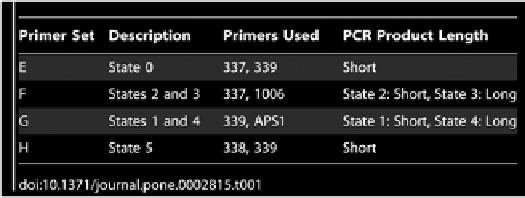
is similar to colony PCR, except the template is not a single colony, but part of the
induced culture, likely containing a mixture of inverted genotypes. Six different
primers in four permutations were used to probe for the presence or the absence
of PCR products (Tables 1, 2 and Fig. 2B). The assay was chosen because, as the
orientation of the DNA fragments change, different permutations of primers will
amplify different length segments or generate no product at all. This method
allowed us to detect very sensitively the presence or absence of a certain orienta-
tion of DNA, based on the presence or absence of PCR amplification product of
different lengths. Culture PCR was much more accurate than detection by fluo-
rescence, as first generation inversion (states 1 and 2) would show no fluorescence
and, it turns out, production of the end states is excessively rare. The sensitive
nature of PCR amplification allowed the detection of rare inversions. The inver-
sion state was also verified by sequencing of the PCR product bands and by direct
sequencing of the vector.
Table 1.
Culture PCR primer sets and lengths of their PCR products. See Fig. 2B for their locations on the
switch.
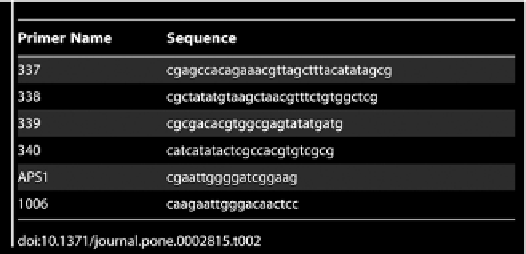
Table 2.
Sequences of primers used to interrogate the inversion products.
Figure 3 shows the results of our experiments. In these experiments cells were
grown in LB at 37°C and exposed to either arabinose or tetracycline. The hin
mirrored pair was replaced with hixC, a variant inversion sequence which allows

Search WWH ::

Custom Search