Biology Reference
In-Depth Information
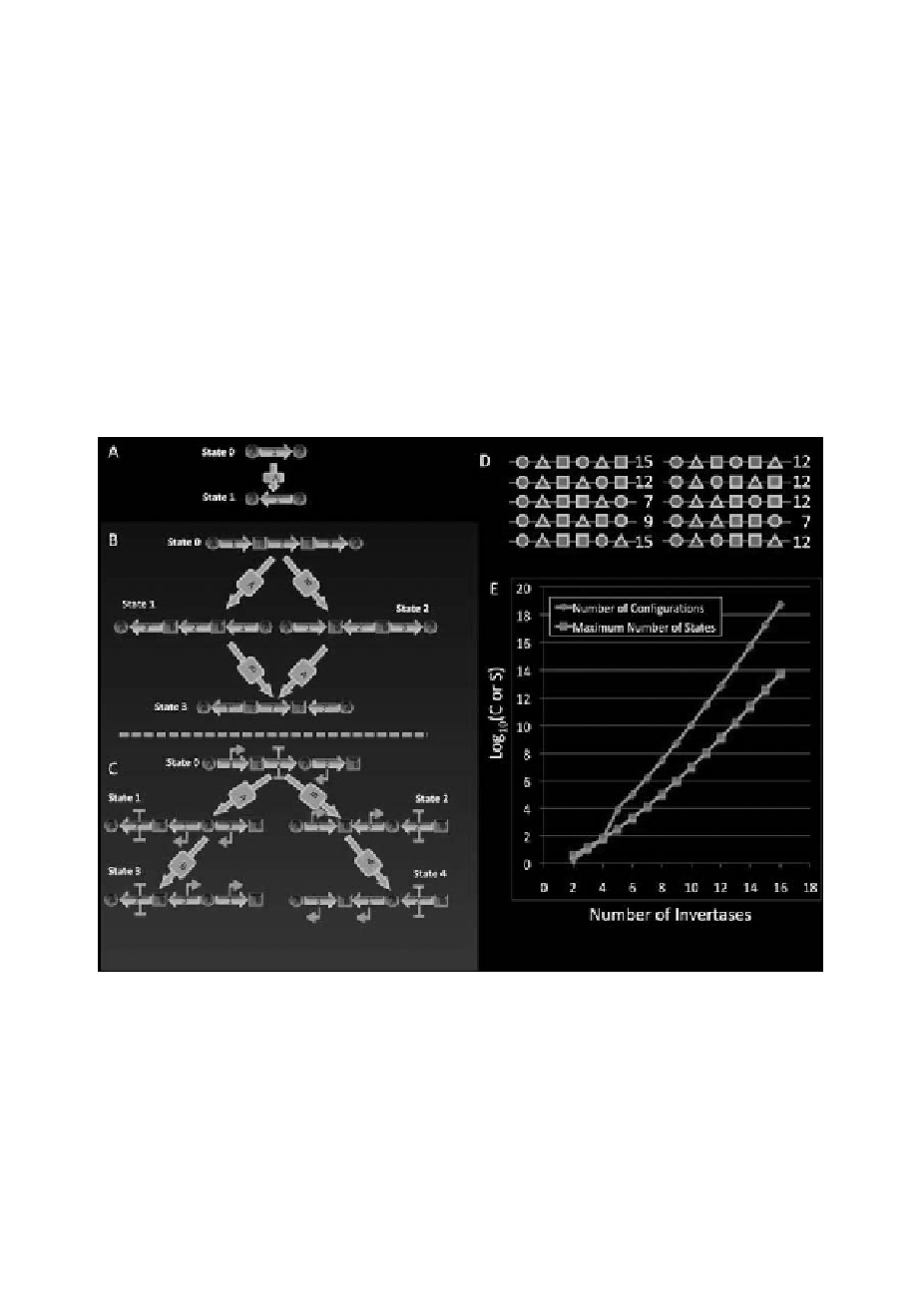
is shown in Figure 1E. Devices accepting input from 10 invertases have maximal
state-spaces of nearly 107.This is a large space made accessible by addition of a
region of DNA on the order of one gene in size (not counting the recombinases).
Of course, not every configuration has this full state space. For example, figure
1C is the only configuration of two pairs of sites that has the full state. The other
configuration (Figure 1B) has a cycle with the final states of the input sequences
{A,B} and {B,A} being identical. Figure 1D shows all the configurations of three
pairs of sites with the number of distinguishable states available to each shown to
its right. Only two of the ten configurations show the full rank state-space. The
others can “remember” only subsets of the input sequences. Nonetheless, as a sys-
tem to remember which of a sequence of N inputs occurred, these types of devices
could be immensely efficient in terms of size and operation.
Figure 1.
How invertase site configuration defines different “machine behaviors”.
A) A single invertase can only
flip one region ON or OFF. B and C) With two invertases there are two possible intercalated configurations
of sites. In B, when one pair of sites is fully contained by another the resultant number of possible “states” of
the system is only four, whereas in C, when the site pairs are staggered, five states are accessible and any input
sequence results in a different output of the system. C also shows a configuration of promoters and terminators
such that states 3 and 4 have promoters pointing outside the machine regions. This configuration is the one
experimentally created. D) Possible configurations of three different pairs of recognition sites and the number of
possible states. E) How both the number of such configurations and the maximal number of states grows with
number of pairs.
Search WWH ::

Custom Search