Information Technology Reference
In-Depth Information
corresponds to the first letter of each word in its entirety. Schwartz[10] proposes
an algorithm using these patterns to recognize abbreviations from biomedical
publications. We improve upon the algorithm of Ariel and found abbreviations
of diseases in abstracts.
3
Information Extraction in HaDextract System
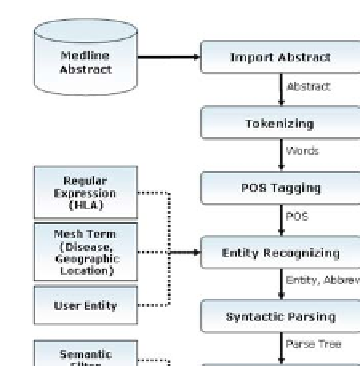
Overall system architecture is displayed in Fig.2. 'Import Abstract Component'
downloaded abstract XML file from PubMed. 'Tokenizing Component' split ab-
stract text into words. 'POS Tagging Component' found POS of each word using
fnTBL POS Tagger offered by Ratnaparkhi[11]. FnTBL was trained with GE-
NIA Corpus 3.0 to search suitable POS in the biomedical domain. 'Entity Rec-
ognizing Component' searched entity names using regular expression and MeSH
keywords, and 'Syntactic Parsing Component' created parse tree using Collins
Parser[12]. 'Semantic Interpret Component' extracted HLA-disease interaction
information using extracted entities, parse trees, relation keywords, and filtering
keywords.
3.1 Relation and Filtering Keyword
In an attempt to find HLA-disease relation information, we developed relation
and filtering keywords determined by domain experts from 309 HLA publications.
Fig. 2.
HaDextract System Architecture




Search WWH ::

Custom Search