Information Technology Reference
In-Depth Information
The new descriptors of evolutionary information, including the mean and standard
deviation of the three biochemical features, were also found to improve classifier
performance. These descriptors indicate how well the biochemical properties of an
amino acid position are conserved in the sequence alignment obtained from PSI-
BLAST search. As shown in Table 2, the use of
H
m
,
K
m
,
H
d
, and
K
d
for input encoding
gave rise to 75.99% prediction strength with 77.70% sensitivity and 74.29% specific-
ity. This classifier achieved similar levels of MCC (0.39) and AUC (0.84) as the RF
constructed using PSSM. The RF classifier was constructed using 1000 decision trees
and
m
= 8. However, adding
M
m
and
M
d
to the input vector did not result in further
improvement of classifier performance (data not shown).
To investigate whether classifier performance could be further improved by combin-
ing the different types of evolutionary information for input encoding, RF classifiers
were constructed using PSSM,
H
m
,
H
d
,
K
m
and
K
d
in addition to the three biochemical
features. Since the input vector had 297 variables (27 inputs for each of the eleven resi-
dues in a data instance), the training parameter
m
was set to
⎣ ⎦
297
= 17 for the forest
with 1000 decision trees. As shown in Table 2, the resulting classifier achieved the
highest level of prediction strength at 78.14% with 78.06% sensitivity and 78.22%
specificity. This RF also had the highest level of MCC (0.43) and ROC AUC (0.86)
among all the classifiers (Table 2). The results suggest that the new descriptors capture
certain evolutionary information that is not contained in PSSM, and thus combining the
different types of evolutionary information for input encoding gives rise to the most
accurate classifier for DNA-binding site prediction.
1
0.8
0.6
0.4
HKM
HKM+EI
0.2
0
0
0.2
0.4
0.6
0.8
1
False positive rate
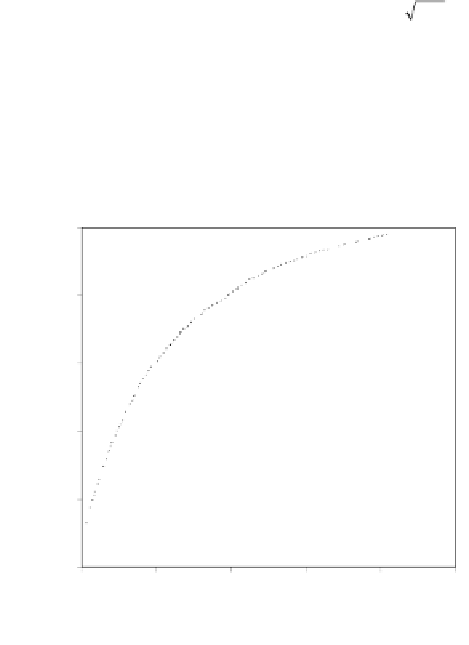
Fig. 1.
ROC analysis to show the effect of evolutionary information on random forest classifi-
ers constructed using the PDNA-62 dataset with the atom distance-based definition of DNA-
binding residues. HKM represents the classifier trained with the three biochemical features (
H
,
K
and
M
), and HKM+EI indicates the classifier using two types of evolutionary information
(PSSM,
H
m
,
H
d
,
K
m
and
K
d
).























Search WWH ::

Custom Search