Database Reference
In-Depth Information
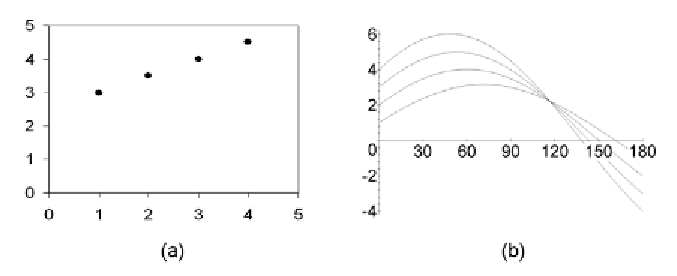
Figure 5. Transformation of the objects into parameter space for correlation clustering as performed
by the algorithm CASH: (a) correlation cluster in original space; (b) representation of the objects as
trigonometric functions in parameter space
arbitrary point of the parameter space. See Figure
5 for an illustration of the transformation to param-
eter space. The algorithm CASH decomposes the
trigonometric functions recursively until a point
in the parameter space is found in which many
trigonometric functions coincide. That means,
even if different planes intersect each other or
are hidden in a noisy environment, the points in
the parameter space representing the correlation
clusters exists and can be found. With increasing
dimensionality the number of required decompo-
sitions of the trigonometric functions increases.
Therefore, also the processing time increases, but
is still of cubic order in the number of dimensions.
Of all algorithms presented in this section, CASH
reveals least sensitivity with respect to noise ob-
jects and intersecting clusters.
ity measures can not satisfactorily represent the
complex human notion of similarity. Consider
for example clustering web pages, which may
contain images, text and hyperlinks. It is difficult
to define an appropriate similarity measure based
on the content of the web pages. As a valuable
source of side information, ratings of users on the
similarity of some selected pages can be integrated
into clustering. As an example from biomedicine,
consider clustering of gene expression data. The
expression levels of thousands of genes can be
measured simultaneously using the micro-array
technology. Clustering is often applied to detect
functionally related genes. There are two major
challenges associated with clustering gene ex-
pression data: First, the curse of dimensionality,
which can be addressed by subspace or projected
clustering. Secondly, there exists a huge amount
of side information on functionally related genes,
where most of this knowledge is contained in
publications available in biomedical literature
databases. Similar to the webpage example, the
side-information is often incomplete, i.e. there are
genes for which we have expression measurements
but no literature information. As demonstrated
in (Zeng et al. 2007), semi-supervised clustering
exploiting both sources of knowledge, the feature
information of gene expression as measured in the
SeMI-SuPervISed cluSterIng
Semi-supervised clustering is an emerging area
which evolved from an important need of numer-
ous applications: integrating side-information
or supervision into clustering. Semi-supervised
clustering may be beneficial for bridging the
so-called semantic gap in human-computer
interaction. Often, purely feature-based similar-
Search WWH ::

Custom Search