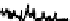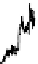Chemistry Reference
In-Depth Information
by peak-to-peak resolution. Additional chromatographic parameters such as peak
width, tailing factors, and column efficiency may also be used.
The parallel study (reference standard and test protein) is also used to visually
compare each peak's relative retention time, response (retention time and area), the
number of peaks, and the overall elution pattern. This comparison is often comple-
mented by mixing the two samples (1:1, v/v) and evaluating the peak response ratios
and elution pattern. If all peaks in this mixed sample have the same relative retention
times and peak response ratios, then the identity of the protein test sample can be
confirmed. Significantly different retention times are also an indication of system
variability, while the appearance of new or broader peaks indicates nonequivalence.
Computer-aided pattern recognition software and other automated approaches
have been used on occasion to examine the degree of difference or similarity when
comparing two different peptide maps, but have not gained routine acceptance.
7.6.3 m
ASS
S
Pectrometry
In
P
ePtIde
m
APPIng
At the Investigational New Drug (IND) phase, limited validation is necessary—
typically, only an approved test procedure that includes system suitability as a test
control. Sometimes termed
qualification,
, complete characterization of the individual
peaks is not needed. As the regulatory process proceeds, a partial validation may be
needed to give assurance that the method performs as intended in the development
of a map for the test protein. However, validation of peptide mapping in support of
further regulatory submissions requires a rigorous characterization of each of the
individual peaks in the map. Methods that are used to characterize the peaks in a
map commonly use mass spectrometry (MS).
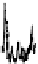
In Figure 7.13, the LC/MS separation of a tryptic digest of alpha-1 acid glyco-
protein is an example of how MS can be used to characterize a peptide map. The
MS detection was performed with a quadrupole time-of-flight (Q-TOF) mass
47.99
100
SIC 657
38.58
43.35
51.23
40.30
35.13
33.50
57.55
62.16
17.83
23.62
26.73
14.18
71.70
0
10.00
20.00
30.00
40.00
50.00
60.00
70.00
Time
FIgure 7.13
LC/MS separation of a tryptic digest of alpha-1 acid glycoprotein. Conditions
are similar to Figure 7.11, except using a longer gradient and formic acid as a modifier. Data is
plotted as a selected ion chromatogram for m/z 657, a signature ion for glycopeptides result-
ing from carbohydrate fragments.