Chemistry Reference
In-Depth Information
been used to digest proteins for peptide mapping, as a way to avoid autodigestion of
the enzymes [39].
7.6.1.2 chromatographic separation
Many different techniques, as well as different modes of chromatography, are used
to separate peptides for mapping. These include various forms of polyacrylamide
gel electrophoresis (PAGE), capillary electrophoresis (CE), and reverse phase high
performance liquid chromatography (RP-LC), ion exchange (IEC), and hydrophobic
interaction chromatography (HIC).
RP-HPLC is arguably the most common technique employed, and the column,
mobile phase, and gradient or (rarely) isocratic conditions used can be critical to the
success of the separation.
Columns used for peptide maps are generally porous silica, 1.7 to 5.0 µm in size,
pore sizes ranging from 100Å to 300Å, with ligands of C
18
(USP column charac-
terization L1) or C
8
(USP column characterization L7). Temperature control of the
column is important for good repeatability. The most common mobile phases used
consist of water and acetonitrile, with various additives, for example, trifluoroacetic
acid (TFA) or formic acid. If a buffer must be used, phosphate buffers provide the
most flexibility for the selection of pH, although some thought must be given to
alternative buffers if MS detection is used. Due to the complexity of the resulting
sample, shallow gradient separations are generally recommended, with segments
sometimes optimized using step functions or different slopes to give better resolution
of important regions. Detection at low UV wavelengths, for example, 200 to 230 nm,
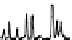
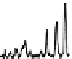
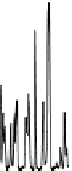
is typically due to limited chromophores. An example chromatogram of a peptide
map is illustrated in Figure 7.12.
0.0
20.0
Time in Min.
FIgure 7.12
Example of a peptide map separation of a phosphorylase digest. A 2.1 by
100 mm 1.7-µm C18 column and a linear gradient of acetonitrile were used, with TFA as a
modifier. Detection was UV at 214 nm.