Biomedical Engineering Reference
In-Depth Information
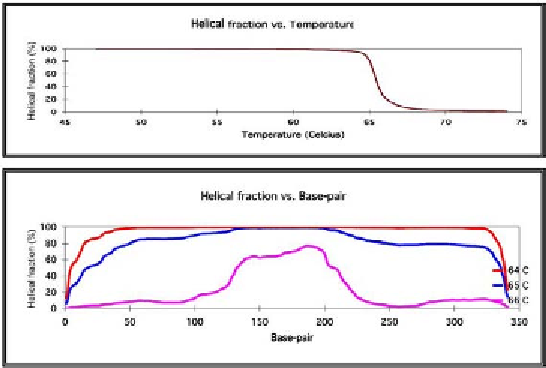
Fig. 2. The melting profile of exon 3 of the
G6PT1
gene. The DNA sequence of the
amplicon is: CTGCCCCATCTGACCCCACCCTCAacatgggcagtaggctggacacctac
gtgtcgccctctgccccacagtggtttgagccatctcagtttggcacttggtgggccatcctgtcaaccagcatgaacctg
gctg
g
agggctgggccctatcctggcaaccatccttgcccagagctacagctggcgcagcacgctggccctatctg
gggcactgtggtgtggttgtctccttcctctgtctcctgctcatccacaatgaacctgctgatgttggact
ccgcaacctggaccccatgccctctgagggcaagaagggtGAGCCCCCACCCAGACCGACCACT.
The uppercase letters are primer sequences. The letter in bold type is the nucleotide
position of the mutation in the wild-type sequence, i.e., 1563G>A. The mutation is
located at nucleotide position 136 of the amplicon. The sequence is shown in the sense
of direction.
2.
PCR amplification (
see
Notes 3
and
4
) is conducted using the primer pairs and
3.2. DHPLC Analysis
DHPLC analysis is typically conducted at temperatures from 51 to 75°C.
The WAVE utility software (Transgenomic Inc.) helps determine the correct
temperature for mutation scanning based on the sequence of the wild-type
DNA. The calculated melting behavior is then used to predict the temperature
suitable for mutation scanning of the fragment. Separation of heteroduplex
DNA from homoduplex DNA depends on a greater proportion of the nonhelical
form in the vicinity of the mismatched bases. To achieve the best possible
separation, the region containing the mutation should have a helical fraction
between 30 and 99%. For example, for DHPLC analysis of exon 3 of the
G6PT1
tridge is packed with C18 alkylated, polystyrene-divinylbenzene polymeric