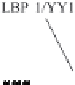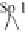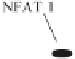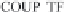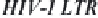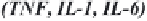Biology Reference
In-Depth Information
cytoplasm of several cell types in physical association with the inhibitory mole-
cule known as I-kB, which prevents its migration into the cell nucleus (Siebenlist
et al., 1994). Several pathways, including activation of protein kinase C, lead to
phosphorylation of I-kB and to the consequent dissociation of phosphorylated
I-kB from NF-kB ( Franzoso et al., 1994). Two binding sites for NF-kB in the
core enhancer region of the HIV-1 long terminal repeat ( LTR) allow binding
of the transcription factor with consequent triggering or potentiation of viral
transcription ( Fig. 14.1). Of note is the fact that di¨erent clades of HIV-1 are
characterized by di¨erent numbers of NF-kB binding sites in the LTR, whereas
HIV-2, and its cognate simian immunode®ciency virus (SIV ), possess only 1 kB
binding region. Clade C HIV-1, rapidly spreading in several regions of sub-
Figure 14.1. Transcriptional control elements of the HIV-1 LTR. The structure of HIV-1 LTR
and the binding sites for cellular transcription factors are shown. Enhancer sequences and a region
of the HIV-1 LTR known as the negative regulatory element (NRE ) as well as the Tat responsive
element (TAR) in the RNA transcripts are shown. COUP-TF, chicken ovalbumin upstream pro-
moter transciption factor; NFAT-1, nuclear factor of activated T cells; GR, glucocorticoid receptor;
USF-1, upstream stimulating factor; EBS, Ets binding factor; T-cell factor (TCF-1), (or lymphoid
enhancer-binding factor, LEF), NF-kB, nuclear factor-kB; AP-1/2, activating protein-1 or 2, com-
posed of heterodimers of Jun and Fos; Sp-1, promoter-speci®c transcription factor; LBP-1, leader
binding protein-1; YY1, ying-yang-1.