Information Technology Reference
In-Depth Information
A
A
B
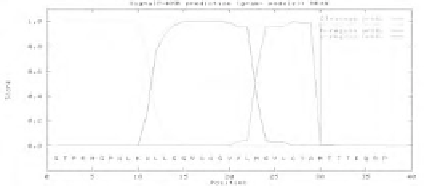
Figure 2.
Prediction of signal peptides for gram-negative bacteria, using NN- and
HMM versions of SignalP. C, S- and Y-scores represent, respectively, the output
from cleavage site networks, the output from signal peptide networks, and the output
of the combined cleavage site score, given by
.
n-region
: positively
charged amino-terminal segment; h-region: central hydrophobic segment; and
c-
region
: polar c-terminal segment.
NrfH
(SignalP-NN) - Positive results; Cut-off
between Ala29 and Met30. NrfH (SignalP-HMM) - Positive results; Cut-off
between Ala29 and Met30.
Y
i
Ci
'
dSi
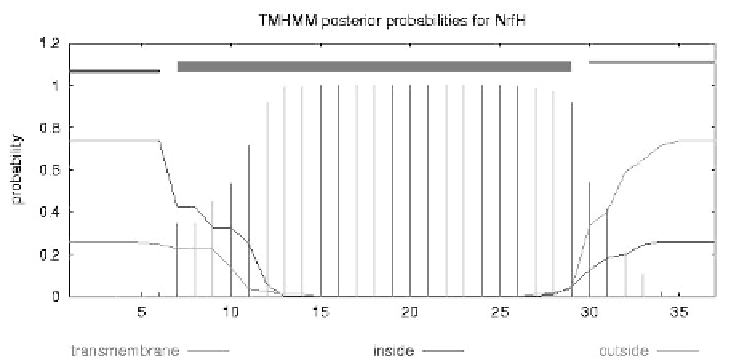
Figure 3.
Transmembrane helix prediction for NrfH subunit by TMHMM 2.0. NrfH
- transmembrane helix: segment 7-29; inside: segment 1-6; and, outside: segment
30-37.
3. Conclusions
The first issue to be considered when predicting of signal peptides and other protein sorting
signals is which program shall be used. The profusion and the continuous rising of protein