Information Technology Reference
In-Depth Information
3.4. Secondary Structure, Solvent Accessibility and Protein Fold Prediction
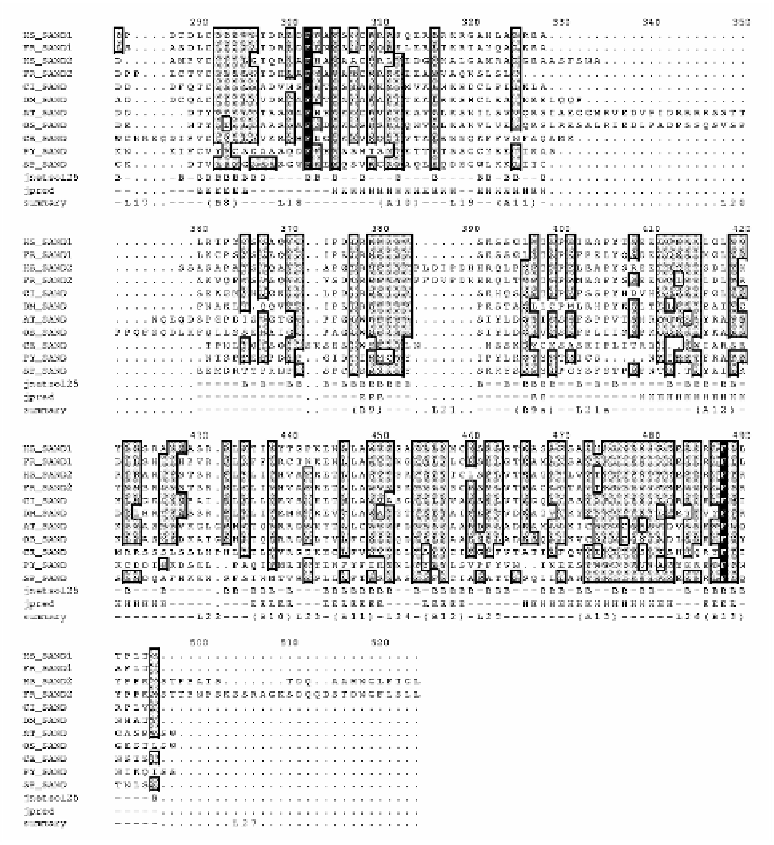
The C-termini of the SAND proteins are predicted to contain fifteen E-strands, thirteen D-
helices and 29 loops. All E-strands are predicted to be largely solvent inaccessible, as are
four D-helices A2, A7, A8 and A9. Eight of the thirteen D-helices display an amphipathic
pattern (A3, A4, A5, A6, A10, A11, A12 and A13). These amphipathic D-helices are likely
to be located on the outer surface of the protein with one side of the D-helix facing the
solvent and the other the hydrophobic interior. The extreme C-termini of the SAND2
sequences are 20-30 residues longer than that of other SANDs (Figure 3) whilst the N-
termini of the SAND1 proteins are 40 residues longer (data not shown).
The C-termini of SAND is likely to contain 3 structural domains. These are possibly
a layered DE-sandwich followed by an D-helical bundle structure and a second layered-DE-
sandwich. It is possible that the first and third domains form a non-contiguous TIM barrel