Information Technology Reference
In-Depth Information
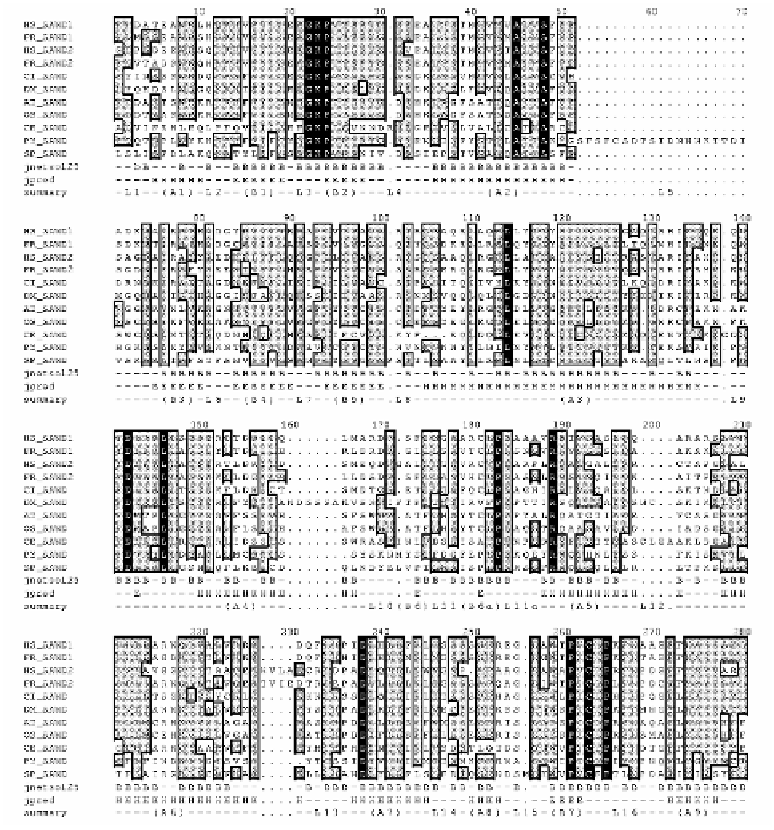
Figure 3.
(continued on the subsequent page) An alignment of eleven
representative C-termini SAND amino acid sequences. Sequence identifiers are
defined in Table 4. The Jpred consensus secondary structure prediction (Jpred) is
supplied where H is D-helix, E is E-strand and - is loop region. A numbering scheme
for the secondary structural elements is provided (Summary) where A is D-helix, B
is E-strand and L is loop. The Jnet side-chain solvent accessibility is predicted
(Jnetsol25) where “B” denotes that 25% or less of the side-chain is solvent
inaccessible and “.” or “-“ denotes that 25% or more of the side-chain is solvent
accessible. Invariant residue positions are formatted with white characters on a black
background. The Figure was generated by the JEMBOSS multiple sequence
alignment editor.