Information Technology Reference
In-Depth Information
(Tables 1-3). The expected accuracy and the strengths and weaknesses of the methods are
highlighted. These methods outlined can be of value in protein structure predictions.
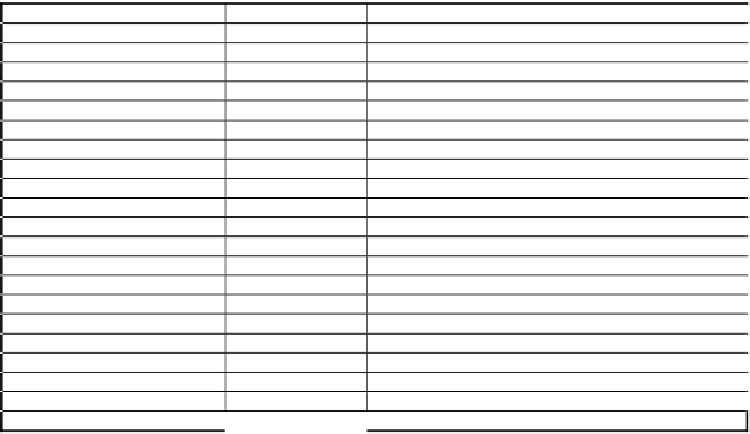
Table 4b.
Seventeen SAND sequences identified in nine vertebrate species.
Organism
SAND identifier
Accession Number
Mammals
Homo sapiens
HS_SAND1
SPTR:Q9BRF3
Homo sapiens
HS_SAND2
SPTR:O94949
Mus musculus
MM_SAND1
SPTR:Q9CYS2
Mus musculus
MM_SAND2
SPTR:Q8BMQ8
Rattus norvegicus
RN_SAND1
REFSEQ:XP_236627
Rattus norvegicus
RN_SAND2
REFSEQ:XP_226493
Macaca fascicularis
MF_SAND1
SPTR:Q95KG9
Birds
Gallus gallus
GG_SAND1
EMBL:GGA395913
Gallus gallus
GG_SAND2
EMBL:BU209213 EMBL:BU258474
Amphibians
Xenopus tropicalis
XT_SAND1
EMBL:AL849442
Xenopus tropicalis
XT_SAND2
EMBL:BQ388616 EMBL:AL779783 EMBL:BJ072986
Fish
Danio rerio
DR_SAND1
EMBL:BX293991
Danio rerio
DR_SAND2
EMBL:BX927379
Fugu rubripes
FR_SAND1
SPTR:Q9YGN1
Fugu rubripes
FR_SAND2
EMBL:CAAB01003001
Tetraodon nigroviridis
TN_SAND1
EMBL:CAF96888
Tetraodon nigroviridis
TN_SAND2
EMBL:CAG07009
2. Material and Methods
2.1. Identification of SAND Homologues
Previously reported SAND protein sequences [25] were used to query public databases
using version 2.2.6 of the NCBI BLAST algorithm [1]. BLASTP was used to search protein
databases with the SAND protein sequences and BLASTX was used to search these
databases with translated SAND nucleotide sequences. Protein databases searched included
SWISSPROT release 42 and SWISSPROT TrEMBL release 25 [30]. Nucleotide databases
searched included EMBL release 77 [31], ENSEMBL release 19 [32] and unfinished
genomic sequences (http://www.ncbi.nlm.nih.gov/BLAST). Translations of these database
sequences were also searched with translations of known SAND sequences using
TBLASTN. Putative SAND gene sequences were verified by comparisons with EST data
using BLASTN.
2.2. BLASTP against NRL3D and Other Protein Sequence Characterisation
An investigation of the SAND sequences was carried out using the web-based analysis tool
PIX (http://www.hgmp.mrc.ac.uk/Registered/Webapp/pix/). PIX helps to identify regions
of interest in a protein sequence. PIX runs several protein analysis programs on a query
sequence and notifies the user via e-mail when the results are ready to be inspected. The