Information Technology Reference
In-Depth Information
A
B
C
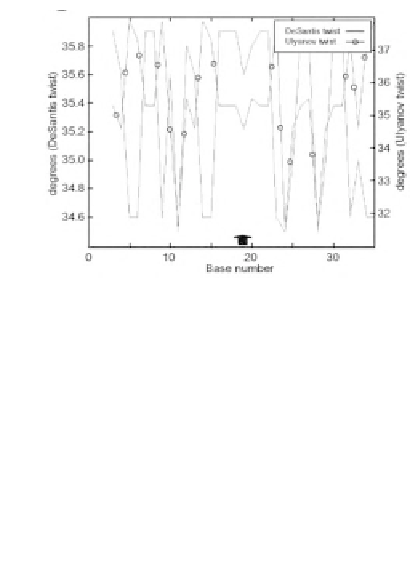
Figure 13
: Analysis of breakpoint sequences in human
chromosomal DNA
A:
Flexibility of DNA obtained
from conformational energy calculations, expressed as
dinucleotide twist, roll and tilt angles. Thick arrow:
breakpoint. (plot.it server)
B.
Twist angles in a
sequence 40 bp, determined from NMR data (empty
circles), and as predicted based on conformational
energy calculations. Thick arrow: breakpoint. (plot.it
server)
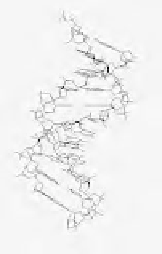
C.
Predicted 3-D model of the short breakpoint
motif CCAGCCTGG, built by the model.it server using
the consensus scalel of DNA curvature, and the raw
models being refined by simulated annealing (model.it
server).
[16] Marini, J.C., et al., A bent helix in kinetoplast DNA. Cold Spring Harb Symp Quant Biol, 1983.
47 Pt
1
: p. 279-83.
[17] Griffith, J., et al., Visualization of the bent helix in kinetoplast DNA by electron microscopy. Cell,
1986.
46
(5): p. 717-24.
[18] Ulanovsky, L., et al., Curved DNA: design, synthesis, and circularization. Proc Natl Acad Sci U S A,
1986.
83
(4): p. 862-6.
[19] Ulanovsky, L.E. and E.N. Trifonov, Estimation of wedge components in curved DNA. Nature, 1987.
326
(6114): p. 720-2.
[20] Diekmann, S., Sequence specificity of curved DNA. Febs Lett, 1986.
195
(1-2): p. 53-6.
[21] Koo, H.S. and D.M. Crothers, Calibration of DNA curvature and a unified description of sequence-
directed bending. Proc Natl Acad Sci U S A, 1988.
85
(6): p. 1763-7.
[22] Haran, T.E., J.D. Kahn, and D.M. Crothers, Sequence elements responsible for DNA curvature. J Mol
Biol, 1994.
244
(2): p. 135-43.
[23] Goodsell, D.S. and R.E. Dickerson, Bending and curvature calculations in B-DNA. Nucleic Acids Res,
1994.
22
(24): p. 5497-503.
[24] http://www.hgmp.mrc.ac.uk/Software/EMBOSS/.
[25] http://www.hgmp.mrc.ac.uk/Software/EMBOSS/interfaces.html.
[26] http://esti.haifa.ac.il/~leon/cgi-bin/curvatur/.
[27] Shpigelman, E.S., E.N. Trifonov, and A. Bolshoy, CURVATURE: software for the analysis of curved
DNA. Comput Appl Biosci, 1993.
9
(4): p. 435-40.
[28] http://www-personal.umich.edu/~mensur/software.html.
[29]
Pedersen, A.G., et al., A DNA structural atlas for Escherichia coli. J Mol Biol, 2000.
299
(4): p. 907-30.
[30]
Jensen, L.J., C. Friis, and D.W. Ussery, Three views of microbial genomes. Res Microbiol, 1999.
150
(9-10): p. 773-7.
[31]
Vlahovicek, K., A. Gabrielian, and S. Pongor, Prediction of bendability and curvature in genomic
DNA. J. Mathematical Modelling and Scientific Computing, 1998.
9
: p. 53-57.
[32]
Ulyanov, N.B. and T.L. James, Statistical analysis of DNA duplex structural features. Methods
Enzymol, 1995.
261
(120): p. 90-120.
[33]
Bolshoy, A., et al., Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles.
Proc Natl Acad Sci U S A, 1991.
88
(6): p. 2312-6.