Biomedical Engineering Reference
In-Depth Information
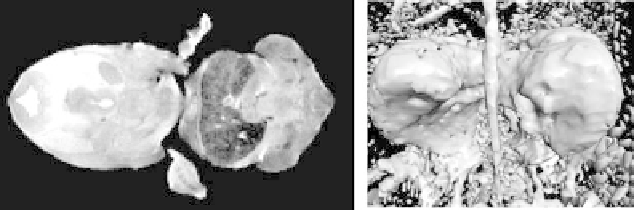
Figure 8.4: (Left) one slice of a 256
×
128
×
128 MR scan of a mouse embryo.
The central dark structure is its liver. (Right) a dual-threshold surface rendering
highlights the segmentation problem.
first involves fitting to discrete edges and then to the gradient magnitude. This
produces a significant improvement over the result in Fig. 8.4. Figure 8.8(a)
presents several other structures that were segmented from the mouse embryo
dataset. The skin (gray) and the liver (blue) were isolated using computational
initialization. The brain ventricles (red) and the eyes (green) were segmented
with interactive initialization.
The same set of initialization and surface deformation steps may be com-
bined to extract a model of a spiny dendrite from the transmission electron
microscopy (TEM) scan presented in Fig. 8.6(a). An isosurface extracted from
the scan is presented in Fig. 8.6(b). Figure 8.7 shows the results of the pro-
posed method compared to the results of a manual segmentation, which took
approximately 10 hours of slice-by-slice hand contouring. The manual method
suffers from slice-wise artifacts, and, because of the size and complexity of the
dataset, the manual segmentation is unable to capture the level of detail that
we obtain with the surface-fitting results. Manual segmentation can, however,
form connections that are not well supported by the data in order to complete
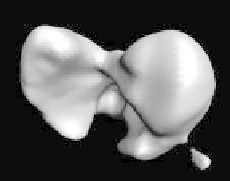
Figure 8.5:
(Left) the initialization of a mouse liver dataset using morphology
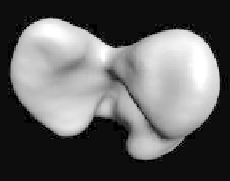
to remove small pieces and holes. (Center) surface fitting to discrete edges.
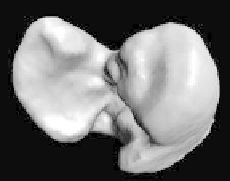
(Right) the final fit to maxima of gradient magnitude.