Information Technology Reference
In-Depth Information
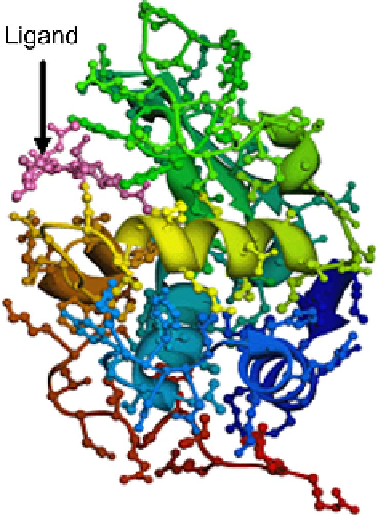
Fig. 1.
(Protein Data Bank code 1HEW). The
protein is colored in rainbow color from one end to the other end. The main chain
is highlighted by ribbons. Side chains extending from the main chain are shown as ball
and stick representations. The magenta colored molecule is the inhibitor ligand, tri-N-
acetylchitotriose. This figure illustrates how a linear protein chain folds up into a three
dimensional structure thereby creating a binding site with which ligand molecules can
interact. All protein figures in this paper have been created using Chimera [4].
An example of a protein: Lysozyme
¯
portant topic in computational biology. Understanding the structure, dynamics
and function of proteins strongly parallels the mapping of words to meaning in
natural language. This analogy is outlined schematically in Fig. 2. The words in
a text document map to a meaning and convey rich information pertaining to
the topic of the document. Similarly, protein sequences also represent the “raw
text” and carry high-level information about the structures, dynamics and func-
tions of proteins. This information can be extracted to obtain an understanding
of the complex interactions of protein within biological systems. Availability of
large amounts of text in digital form has led to the convergence of linguistics
with computational science, and has resulted in applications such as information
retrieval, document summarization and machine translation. Thus, even though
computational language understanding is not yet a reality, data availability has
allowed us to obtain practical solutions that have a large impact on our lives. In
direct analogy, transformation of biology by data availability opened the door
to convergence with computer science and information technology.
Many of the hallmarks of statistical analysis of biological sequences are sim-
ilar to those of human languages. (i) Large data bodies need to be analyzed
Search WWH ::

Custom Search