Biology Reference
In-Depth Information
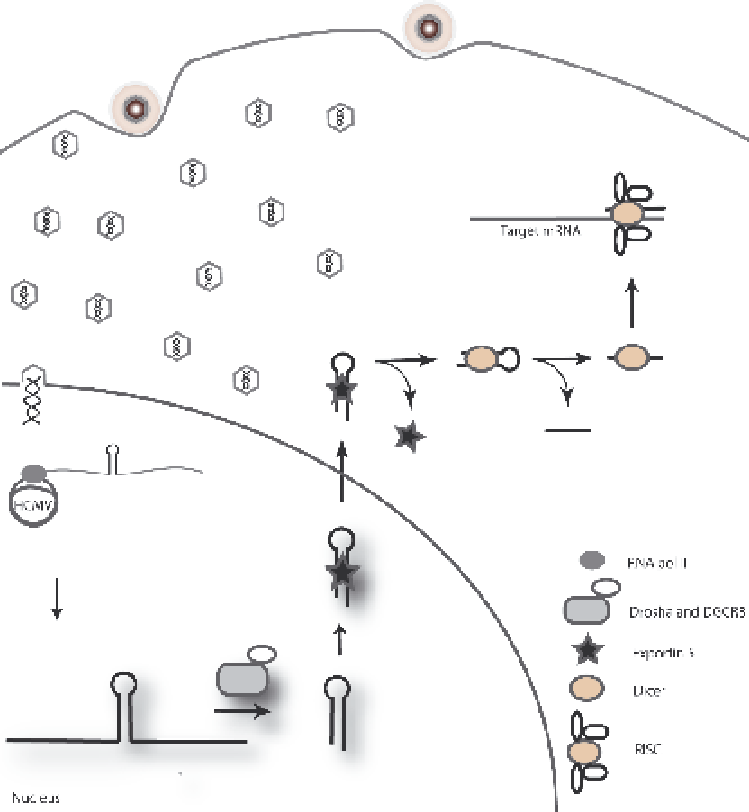
Fig. 2
Biogenesis of miRNAs. Transcription of viral pri-miRNA occurs most often by RNA
polymerase II. Pri-miRNAs are then capped and polyadenylated. Nuclear RNase type III endonu-
clease Drosha then processes the pri-miRNA to pre-miRNA. Pre-miRNAs contain 2-nt 3′ over-
hangs characteristic of processing by RNase type III nucleases. Pre-miRNA is then exported via
exportin-5 from the nucleus. Cytoplasmic type III endonuclease Dicer then cleaves the stem loop
of the pre-miRNA to yield a 22-nt dsRNA. The strand with lowest thermodynamic stability at its
5′ end is chosen as the guide strand and incorporated into the RISC complex. The strand not cho-
sen for incorporation is typically degraded. Guide strand miRNA enables recognition of target
mRNA by RISC. Target mRNAs are then either cleaved or translation is inhibited
(Pfeffer et al. 2005). As such, the reader is encouraged to pursue the topics
included in this section with the host-microbe interaction in mind.
The miRNA transcriptional unit is termed a primary miRNA (pri-miRNA). The
pri-miRNA may possess multiple hairpins or single hairpins (Kim 2005). While
most miRNAs are located within an intron of a protein-coding transcript, miRNAs
can be found in the exons as well as the protein coding potential of the transcriptional