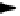Biology Reference
In-Depth Information
Structural Observations in the First Hours After CMV
Infection and Their Limits in Interpretative Value
The last physical hurdle for the infecting viral genome appears to be the nuclear
pore complex. How viral DNA enters the host nucleus is not entirely clear, nor is
it clear how it is protected from degradation by endonucleases once released from
the capsid. One must assume that the viral DNA is first neutralized before or as it
enters the host nucleus. This could be mediated either by the host's histones -
specifically during S-phase when free histones are abundant - forming an easily
silenced chromatin package or by positively charged polyamines neutralizing the
virus DNA's negative charge, as has been reported for HSV1 (Gibson and
Roizman 1971). Spermidine may still be associated with the viral genome when it
leaves the capsid and enters the host nucleus. Because the nucleus is a highly
structured environment, viral genomes are excluded from certain domains, such as
the tightly packed nucleolus. Any observed nonrandom distribution of viral
genomes in the nucleus may therefore be due to exclusion.
We do not know the physical dimensions of the large viral genome of CMV in
the nucleus. A tightly coiled, chromatinized genome may be slightly larger than
an encapsidated genome. Nor do we know whether the viral genome can move
through the nucleus, either by passive diffusion or by active transport. This ques-
tion has been difficult to address, because we cannot see the virus in real time and
must therefore construct a likely sequence of events from observations of fixed
material. The large size of CMV genomes aids microscopic identification and
localization by in situ hybridization, which allows individual viral genomes to be
visualized (Ishov et al. 1997). The size of these signals is close to that obtained
by imaging tegument proteins like pp71 (Fig. 1a and b), i.e., the diffraction point
size, which is limited by the wavelength of light. Signals can only become weaker
when originating from smaller sources. Comparison of encapsidated viruses and
in situ hybridization signals representing CMV genomes suggests that the infected
host nucleus contains genomes that are highly condensed (Ishov et al. 1997). One
caveat is that the completely extended genome (78 µm) would not be visible using
this technique, because signals from any point along the approximately 260-Mb
genomes would be too weak to register with our current techniques. Therefore,
the detectable size, as estimated from what is visible, is equal to or smaller than
the wavelength of light (~300 nm).
Fig. 1
continued) early transcripts (
green
), for IE2 (
blue
) and PML (
red
). IE2 is located like a col-
lar around the emerging transcripts.
d
3T3 cell infected with MCMV (24 h p.i.). The cell is labeled
with antibodies to the 112/113 gene product (
red
) and for viral genomes by in situ hybridization
(
green
). Viral genomes are seen on the outside of the 112/113 labeled prereplication domains.
e
HFF
3 h p.i. by HCMV. Cell is triple-labeled for viral immediate early transcripts (
green
), for the splicing
compartment delineating SC35 (
blue
) and the PML defining ND10 (
red
). The nucleus is outlined in
blue
.
f
MCMV infected 3T3 cells (24 h p.i.). Cells were probed for viral DNA by in situ hybridiza-
tion (
green
) and for the 112/113 gene product by antibodies (
red
). The
hollow red spheres
inside the
nucleus represent the replication compartments. The nucleus is outlined in
blue