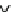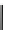Biology Reference
In-Depth Information
Sample A
Sample B
Sample C
Sample D
Reducon
Alkylaon
Protease digeson
Reducon
Alkylaon
Protease digeson
Reducon
Alkylaon
Protease digeson
Reducon
Alkylaon
Protease digeson
iTRAQ
114 -reagent
labeling
iTRAQ
115 -reagent
labeling
iTRAQ
116 -reagent
labeling
iTRAQ
117-reagent
labeling
mix
Caon-exchange column
treatment
Desalng
MS/MS analysis
Fig.
2
Flowchart of the quantitative proteomic analysis with iTRAQ labeling
1. All reagents and buffers are contained in the iTRAQ Reagents
kit.
2. Add 2
L of reducing reagent to each protein sample and incu-
bate for 1 h at 37 °C (reduction) (
see
Notes 22
and
23
).
3. After spin-down, add 1
μ
L of cystein blocking reagent and
incubate for 10 min at room temperature (alkylation).
4. After spin-down, add 10
μ
μ
L of trypsin (0.5
μ
g/
μ
L) and incu-
bate overnight at 37 °C (protease digestion).
5. Bring each vial of iTRAQ reagents to room temperature. Add
70
L of ethanol to each iTRAQ reagent and vortex for 1 min.
After spin-down, transfer the contents of one iTRAQ reagent
vial to one sample tube. Mix and incubate for 1 h at room
temperature.
6. After adding 900
μ
L of LC/MS-grade water, combine the
iTRAQ-labeled peptide samples into one new tube.
μ
To increase the effi ciency of protein identifi cation in shotgun
proteomics analysis using an LC-MS/MS system, the mixture of
iTRAQ-labeled peptides is separated into several fractions using
iCAT Cation Exchange column [
11
].
3.6 Cation Exchange
Column Treatment
1. Check the pH of the mixture of iTRAQ-labeled peptides using
a pH test paper. The desired pH is between 2.5 and 3.5. If the
pH exceeds 3.5, add a small amount of FA for adjustment.