Biology Reference
In-Depth Information
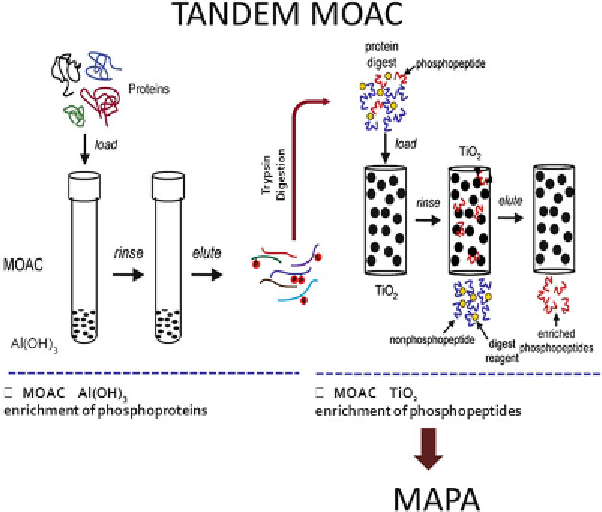
Fig.
4
TANDEM MOAC procedure combined with MAPA [
14
]. Phosphoproteins are enriched fi rst using alumina-
based MOAC [
17
,
18
]. The enriched phosphoprotein fraction is digested and phosphopeptides are enriched
using TiO
2
-MOAC. This phospho-peptide fraction is analyzed by LC-MS and MAPA and subsequently with
multivariate statistics such as PCA or ICA using COVAIN to identify changed abundances of specifi c phosphory-
lation sites in proteins [
12
]
protein kinase (MPK) signaling pathway [
14
]. An essential
requirement for the confi dential identifi cation of novel protein
kinase targets as a regulatory response to environmental pertur-
bations is the accurate quantifi cation of the phosphoproteins/
phosphopeptides. The reason is that protein phosphorylation—
though low abundant—is prevalent and distinguishing changes
is only possible with quantifi cation of the respective phosphory-
lation site of the protein as a result of a switched-on signaling
cascade. Therefore we combined MOAC with MAPA as a quan-
titative phosphoproteomics approach to identify novel protein
kinase targets switched on in signaling processes. As discussed
above the tandem MOAC procedure was combined with MAPA
to identify new MPK substrates [
14
]. In another study novel
phosphoproteins were identifi ed as a response to phytohormone
signaling using the combination of MOAC and MAPA [
12
].
Another study used MOAC and MAPA for the identifi cation of
phosphoproteins during pollen tube growth [
13
].