Biology Reference
In-Depth Information
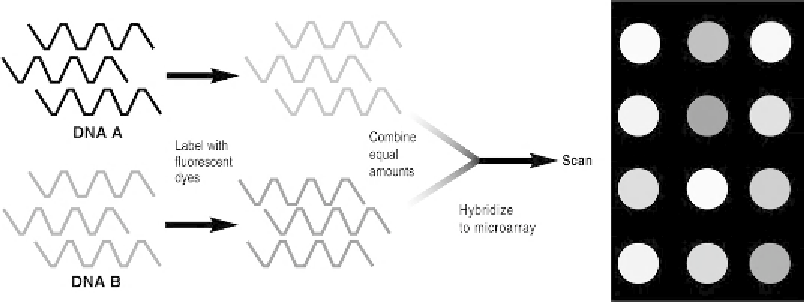
Figure 13.1
In some microarray experiments, cDNAs copied from the messenger RNAs of two
different insects (A and B) are each labeled with a different colored fluorescent dye. The labeled
cDNAs are then combined in equal proportions and used as probes to hybridize with known DNA
or RNA sequences on the microarray. After hybridization, the microarray is scanned, and the colors
and their relative intensities are recorded. If, for example, cDNA from insect A is labeled with red
and cDNA from insect B is labeled with green, when equal amounts of A and B cDNAs are applied
to the array the resulting color patterns indicate which genes are active in insect A and B. A red dot
will indicate high levels of expression of the corresponding gene on the microarray in insect A (and
no expression in insect B). A green dot in that area indicates high levels of expression of that gene
in insect B (and no expression in A). If the same gene is equally active in both insects, then the color
will be a blend of red plus green, or yellow. [The dots on the right side of the illustration are shown
in shades of gray here, but normally would be in color.]
cost of microarrays is declining rapidly as more core facilities are devoted to this
technology and as more arthropod genomes are sequenced.
Analysis of microarray data must deal with the challenge of comprehending
and interpreting the resulting massive amounts of data. As with any new tech-
nology, quality control and adequate statistical methods must be developed and
used (
Kerr and Churchill 2001a,b, Quackenbush 2001
).
13.5.7 Microsatellites
DNA fingerprinting may involve the use of “microsatellite” or “minisatellite”
sequences, which consist of arrays of up to several hundred simple sequence
repeats (SSRs). In arthropods, these repeats most often consist of repeats of
dinucleotides (AC, AT, AG), trinucleotides (AGC, AAC, AAT) or tetranucleotides
(ACAT, AAAT, AAAC) (
Toth et al. 2000
). Microsatellites typically are scattered
throughout the chromosomes of most organisms in both protein-coding and
noncoding regions (
Bruford et al. 1992, Estoup et al. 1993, Ashley and Dow