Biology Reference
In-Depth Information
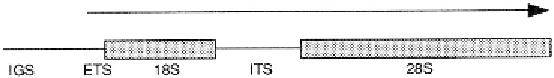
Figure 12.2
A simplified diagram of the ribosomal DNA repeat unit of eukaryotes. IGS is the inter-
genic spacer region. ETS is the external transcribed spacer, 28S is the large subunit rRNA gene. The
arrow indicates the direction of transcription. Most insects have hundreds of ribosomal RNA genes
in tandem array. Some contain R1 and R2 retrotransposable elements (not shown) in specific loca-
tions. Genes with R1 and R2 elements produce nonfunctional product.
rRNA coding region, and an
InterGenic nontranscribed Spacer segment
(
IGS
)
(
Figure 12.2
). Different portions of the repeated transcription unit evolve at dif-
ferent rates. In general, a higher degree of polymorphism has been found in the
noncoding segments
(ETS, ITS, and IGS). The most variable part of the repeated
unit is the intergenic spacer (IGS); it typically contains reiterated subrepeats
ranging from about 50 to several hundred bp in length (
Cross and Dover 1987
).
The coding regions of the repeated unit change relatively little, and can be used
for systematic studies of higher-level taxa or for ancient lineages. Highly con-
served regions are no doubt important for maintaining the characteristic sec-
ondary and tertiary structure of rRNA molecules (
Simon et al. 1991, Van de Peer
et al. 1993, Caterino et al. 2000
).
Ribosomal RNA genes undergo concerted evolution so that the sequence
similarity of members of an rRNA family is expected to be greater within a spe-
cies than between species. Unequal crossing over, gene conversion, and illegit-
imate recombination are responsible for concerted evolution. Ribosomal gene
families are considered to be “quite uniform” (
Ohta 2000a
). Two retrotranspo-
sons, called RI and R2, have been found in the 28S rRNA genes of most insects
(
Eickbush 2002
). These elements have been associated with arthropods for
>
500
million years, and they usually are located at the same position within the 28S
gene. Most R2 elements are located
≈
74bp upstream from the site of RI inser-
tions. R1 and R2 elements lack long terminal repeats (LTRs) and block the pro-
duction of functional rRNA (
Eickbush 2002
). The insect survives because it
contains hundreds of rRNA genes and the R2 elements are kept from invading
too many of them by unknown mechanisms. Surprisingly, most R1 and R2 ele-
ments have not accumulated mutations that would make them inactive. Some
species have more than one family of R1 or R2 elements, and sequence identity
between the different families can be low, suggesting either that each insertion
family is able to maintain its copy number without eliminating other families or
that there has been horizontal transfer of R1 and R2 elements between species.
A phylogenetic analysis of R2 elements and arthropods suggests that multiple