Biology Reference
In-Depth Information
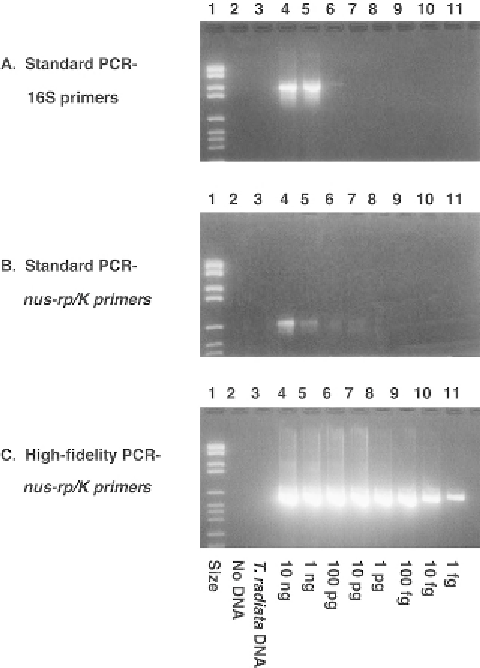
Figure 8.4
Long (or high-fidelity) PCR is more sensitive by
≈
6 orders of magnitude than standard,
allele-specific PCR when microbial DNA is mixed with insect DNA (
Hoy et al. 2001
). The same tem-
plate DNA (a plasmid containing both the
nus
G-
rp
/K and 16S sequences) from the greening bac-
terium was serially diluted from 10 nanograms (ng) to 1 fentogram (fg) and added to 10ng of
parasitoid,
Tamarixia radiata
, DNA. A) 16S primers were used in a standard, allele-specific PCR pro-
tocol. Detectable products are in lanes 4 and 5 only. B)
nus
G-
rp
/K primers were used in a standard,
allele-specific PCR protocol. Weak products are seen in lanes 4 and 5. C)
nus
G-
rp
/K primers were used
with a Long PCR protocol. Strong products were produced in lanes 4-11. Lane 1, DNA size marker;
lane 2, no template-DNA control; lane 3, 10 ng of
T. radiata
DNA only control; lanes 3-11 contain
T.
radiata
DNA
+
10 ng, 1 ng, 100 pg, 10 pg, 1 pg, 100 fg, 10 fg, and 1 fg of the plasmid DNA, respectively.
8.4.9 Multiplex PCR
More than one pair of primers can be used to amplify multiple PCR products
(
Sambrook and Russell 2001, Hamiduzzaman et al. 2010, Meeus et al. 2010
,
Ravikumar et al. 2011
). In
multiplex PCR
, the goal is to amplify several segments