Biology Reference
In-Depth Information
7.3 The Dideoxy or Chain-Termination (Sanger) Method
In brief, the
dideoxy
or
chain-termination
method developed by Sanger involves
de novo
synthesis of a series of labeled DNA fragments from a single-stranded
(ss) DNA template. The ss DNA segment to be sequenced serves as the template
for the synthesis, by base pairing, of a new labeled strand of DNA. Labeling ini-
tially was achieved by labeling with
32
P or
35
S.
Because DNA synthesis is used, the sequencing reaction requires a DNA poly-
merase, labeled deoxyribonucleotides (dNTPs), a primer, and dideoxyribonucle-
otides (ddNTPs) (
Figure 7.1
). Several different DNA polymerases could be used
(Klenow fragment, Sequenase 2, or thermophilic DNA polymerases such as
Taq
),
with different protocols (
Sambrook and Russell 2001
). The dNTPs can be labeled
either with
32
P or
35
S, but
35
S labeling produces sharper bands and improves the
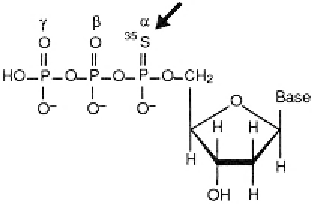
resolution of the autoradiogram. The structure of a dNTP that has been labeled
with
35
S is shown in
Figure 7.1
.
DNA sequencing kits that contain the necessary enzymes and components
can be purchased. Each has specific protocols provided and is ideal for first-time
sequencers, although they are too expensive for large-scale projects (
Sambrook
and Russell 2001
).
The dideoxy or chain-terminating reaction begins by adding a short oligonu-
cleotide primer that is complementary to a region of DNA adjacent to the DNA
segment to be sequenced (
Table 7.1
,
Figure 7.2
). The primer is normally 15-30
nucleotides and is annealed to the template in a preincubation step. Four sep-
arate reactions are set up to determine the position, respectively, of the A, T,
G, and C bases in the template DNA. Each reaction requires a mixture of DNA
polymerase, primers, dNTPs, ddNTPs, and the template DNA. The ddNTPs are
Figure 7.1
Structure of
α
-
35
S-deoxynucleoside triphosphate. Labeling with
35
S results in sharp bands,
which increases the resolution of the sequencing gels.