Biology Reference
In-Depth Information
denatured by an alkali treatment and then brought to a neutral pH. After dena-
turation, the now single-stranded DNA on the filter is incubated with a probe.
The probe base-pairs with the specific nucleotide sequence from the gene of
interest, but not with DNA from plaques containing other genes.
The position of the probe that is hybridized to the immobilized DNA on the
filter can be located by autoradiography. The filter and the original agar sub-
strate are then compared using the marker, and the corresponding plaque is
located on the original agar substrate. A few phage can be picked from each
plaque that yielded a spot on the X-ray film. The phage from that plaque is
used to infect individual new
E. coli
cultures to produce multiple copies of that
phage.
Plaque hybridization allows several hundred thousand plaques to be screened
at once, so a single-copy gene can be isolated from thousands of clones. Because
the DNA that was inserted into the
λ
vector was cut at random, it is likely that
more than one clone (plaque) will contain the DNA of interest. Ideally, at least
one clone isolated by the probe will contain the complete gene, but this out-
come can only be determined after the DNA has been sequenced, a technique
that is described in Chapter 7.
Another technique used to identify specific DNA sequences is called
chromo-
some walking
(
Figure 6.11
). It is particularly useful with
Drosophila
and less useful
with insects for which less genetic information is available. Chromosome walking
is used to isolate a gene of interest for which no probe is available. The gene of
interest
must
be linked to a marker gene that has been identified and cloned.
This marker gene is used as a probe to screen a genomic library. All fragments
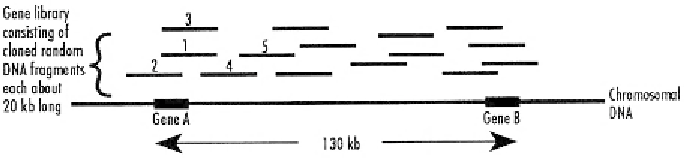
Figure 6.11
Chromosome walking is used to identify a gene of interest when a probe is not avail-
able. It can only be carried out when it is known that the target gene is linked to another gene that
has been cloned and sequenced. First a clone that contains gene A is isolated in fragment 1. This
fragment is sequenced and new probes are synthesized that contain sequences from each end of the
fragment. The new probes are used to identify overlapping DNA clones in the library on each side
of fragment 1, i.e., clones 2, 3, and 4. Clone 4 can, in turn, be sequenced, and used as a probe to
identify clone 5, and so on until gene B is reached.