Biology Reference
In-Depth Information
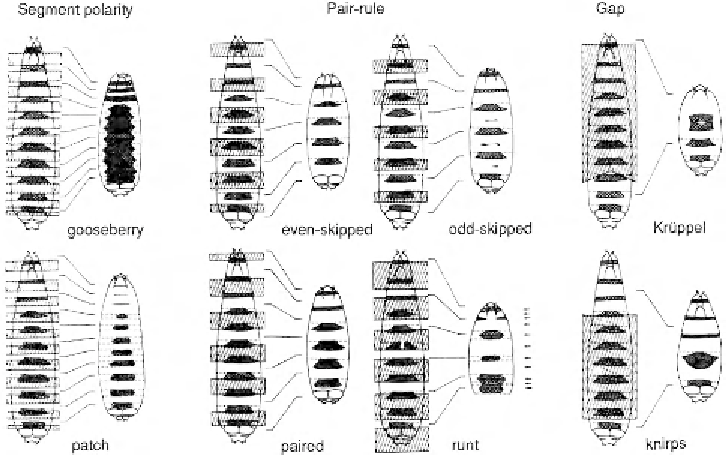
Figure 4.7
Embryonic segment pattern defects are illustrated with selected mutants:
gooseberry
and
patch
are segment-polarity mutants;
even-skipped
,
odd-skipped
,
paired
and
runt
are pair-rule mutants;
and
Krüppel
and
knirps
are gap genes. Dotted regions represent denticle bands on the developing
embryo and dotted lines show the boundaries of segments. Hatched regions indicate the parts
of the pattern that are missing in the mutants. Transverse lines link the corresponding regions in
mutant and wild-type embryos. Arrows indicate where lines of polarity have been reversed.
(From
Nature
287
: 796, 1980.)
4.14.2.1 Gap Genes
Gap genes
were so named because large areas of the normal cuticular pattern
are deleted in individuals with mutant phenotypes (
Figure 4.7
). The three wild-
type versions of the gap genes
Krüppel
+
,
hunchback
+
, and
knirps
+
regional-
ize the embryo by delimiting domains of homeotic gene expression and affect
position-specific regulation of the pair-rule genes
runt
+
,
fushi tarazu
+
,
even
skipped
+
,
paired
+
, and
odd-paired
+
. All three gap-gene products contain DNA-
binding domains.
The gap genes interact to produce sharp boundaries. The establishment of
stable domains by the gap genes is a two-step process: 1) a differential response
to graded levels of maternal determinants, 2) followed by mutual repression
leading to the generation of stable boundaries between adjacent domains. Gap
genes regionalize the embryo by delimiting the domains of later homeotic-gene
expression and this results in position-specific regulation of the pair-rule class
of genes.