Biology Reference
In-Depth Information
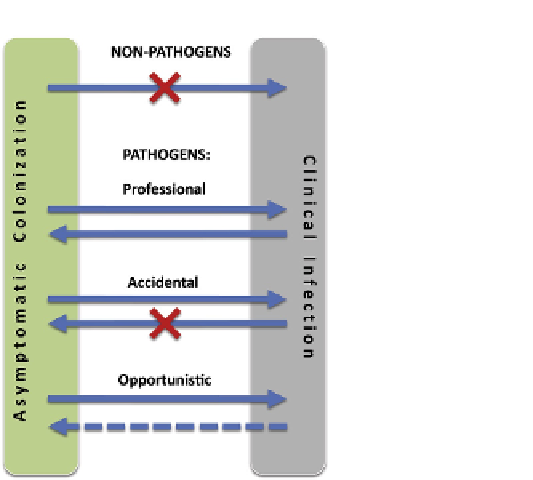
FIGURE 3.3
Different evolutionary types of pathogenic
E. coli
from the perspective of population
dynamics.
against this scenario (
Chattopadhyay et al., 2012
). Then, the derived nature of
pathogens is obvious from the fact that pathogenic isolates (like
Shigella
) tend
to lose or inactivate certain core genes that are evolutionarily original to the
species (
Maurelli, 2007
). Also, pathoadaptive point mutations are of a derived
nature based on the phylogenetic analysis, with the non-pathogens inclined
to have an original (ancestral) variant of the gene (
Sokurenko et al., 2004
;
Weissman et al., 2007
).
A good illustration of the non-pathogen-to-pathogen evolution is pro-
vided by
Shigella
.
Shigella
were originally grouped into four separate species
(
S. boydii
,
S. dysenteriae
,
S. flexneri
,
S. sonnei
), but later it was determined that
E. coli
and
Shigella
should be considered as a single species (
Jin et al., 2002
).
Shigella
/EIEC genomes are colinear to
E. coli
with >90% homologous genes
(
Jin, et al., 2002
). While
Shigella
strains belong to either B1 or ABD phyloge-
netic ECOR groups of
E. coli
(
Figure 3.1
), a more detailed phylogenetic analy-
sis showed their independent emergence from separate
E. coli
lineages (
Pupo
et al., 1997, 2000
). Genetic studies revealed that it is horizontal transfer of clus-
tered virulence genes with low G+C content compared to the rest of the plasmid
that occurred multiple times, each of which gave birth to a new
Shigella
clone
(
Pupo et al., 2000
). Analysis of the region encompassing
cadA
, the antivirulence
gene, from four
Shigella
species demonstrated distinct genetic rearrangements
in each of the regions, again indicating independent origin of different
Shigella
lin-
eages (
Day et al., 2001
). Interestingly, however,
Shigella
lineages emerged only
within one major phylogenetic group of
E. coli
, suggesting that the acquisition
Search WWH ::

Custom Search