Information Technology Reference
In-Depth Information
−
0.04
−
0.02
0
0.02
0.04
0.06
11
1sd
0.08
0.1
0.12
0.14
0.16
0.18
0.2
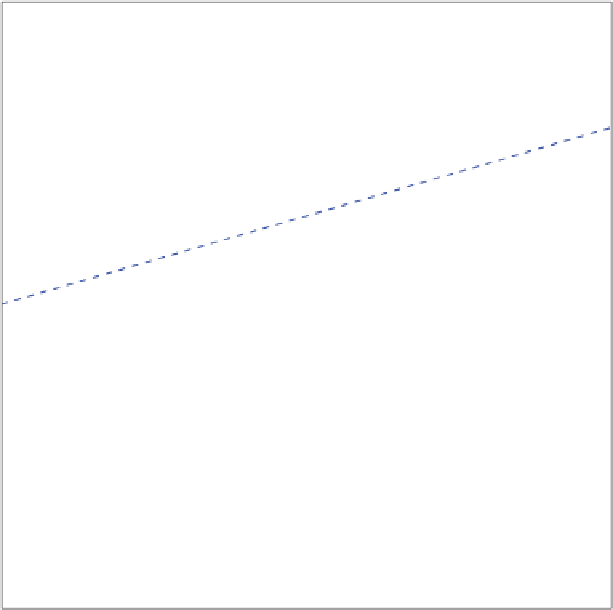
Figure 2.10
Calibrating a biplot axis in the case of normalized data. Biplot drawn by
1
/
2
1
/
2
plotting
(
U
)
2
versus
(
V
)
2
.
Secondly, obtain the corresponding representations on the biplot scaffolding using the
inverse relationship
>
markers.v <- exp(markers.x - 5)
Now apply (2.10) to find the coordinates of
markers.x
in terms of the biplot scaf-
folding:
>
coords.horizontal <- (markers.v / sum(V[1,1:2]^2)) * V[1,1]
>
coords.vertical <- (markers.v / sum(V[1,1:2]^2)) * V[1,2]
Plot
coords.vertical
versus
coords.horizontal
and label with
markers.x
.
The R code for constructing Figures 2.9 and 2.10 is as follows:
function (X = ReactionKinetic.data[,1:4], add = c(2,2),
exp.factor = 0.8, shift = -2, lambda = 1, n.int = 5)
{
options(pty = "s")
par(mar = c(3,3,3,3))
# obtain biplot scaffolding
X.scaled.svd <- svd(scale(X, scale = TRUE, center = TRUE))
col.means <- apply(X,2,mean)
sds <- apply(X,2,function(x) sqrt(var(x)))
USigma <- X.scaled.svd$u %*% diag(X.scaled.svd$d)[,1:2]*lambda