Biology Reference
In-Depth Information
depending whether the data division is represen-
tative. Hence, strati
features is probably the major bottleneck of
metabolomic analyses, particularly when
analyzing human samples in which the entire
study is conventionally achieved at the analyt-
ical level. On the other hand, in plant metabolo-
mics, the ability to isolate compounds makes
NMR experiments then possible.
Metabolites lack the characteristic of sequence
that is central for the identi
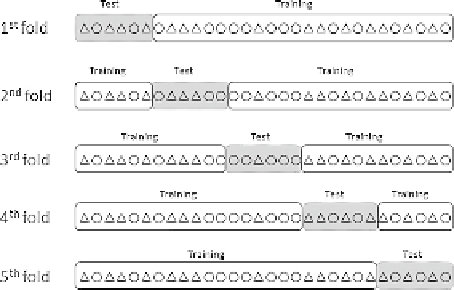
ed resampling is usually
done to guarantee an appropriate representation
of each class in both the training and the test set,
instead of pure random sampling. A
fivefold
cross-validation scheme is illustrated in
Figure 6
.
When a parameter, such as the model
s size,
must be tuned during cross-validation, a nested
procedure is usually desirable. An inner cross-
validation loop is employed for parameter eval-
uation and an outer loop for error estimation.
Parameter tuning must therefore be considered
as an integral part of the training phase, but
such a process is computationally demanding.
'
cation of proteins or
nucleic acids from untargeted analyses. In that
context, MS constitutes one of the key technolo-
gies for the identi
cation of such small mole-
cules. It is usually achieved by comparing
experimental results with standards to relate
measured
m/z
signals to chemical entities.
Several pieces of information can be compared
to reference entries of metabolite databases. It
includes accurate molecular mass, leading to
putative elemental composition and speci
Metabolite Identi
cation: Biological
Validation
The analysis of global metabolic
c
mass spectra such as true isotope patterns. In
addition to the information provided by the
analysis of molecular ions with high-resolution
analyzers,
fingerprints
without prior identi
cation of the variables can
lead to relevant classi
cation models, provided
that proper modeling and validation have been
achieved. These models are useful to distinguish
characteristic metabolic signatures for given
groups of observations and predict reliably
new samples. Metabolite identi
cation process can be
greatly enhanced by the use of fragmentation
patterns. Several levels of reliability have been
proposed by the Metabolomics Standards Initia-
tive (MSI) for the identi
the identi
cation remains
mandatory to validate the model in a biological
context. The identi
cation of unknown
compounds,
115
including (1) an indisputable
cation of discriminant mass
identi
cation based on measurements of stan-
dards in the same experimental conditions, (2)
putatively annotated compounds based on the
comparison with literature data, (3) putatively
characterized compound classes using character-
istic physicochemical properties or spectral simi-
larity, and (4) unidenti
ed compounds.
116
CONCLUSIONS
Appropriate handling of complex MS-based
data structures is required to extract information
hidden within the large amounts of signals
recorded by modern analytical devices. Data
handling is therefore an essential part of the
FIGURE
6
Example
of
fivefold
cross-validation
procedure.



Search WWH ::

Custom Search