Biology Reference
In-Depth Information
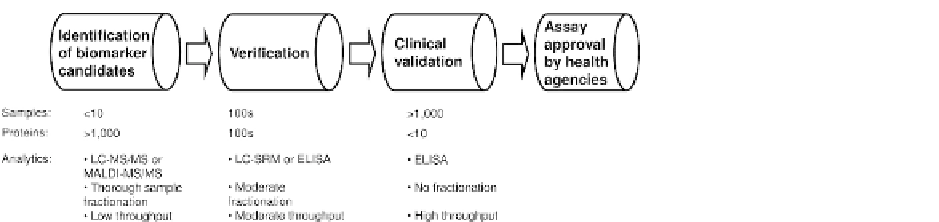
FIGURE 1
The proteomic biomarker
development pipeline. As biomarker
candidates proceed through the pipeline,
the number of clinical samples increases,
while analytical technologies change
from complex and low-throughput mass
spectrometry methods to straightfor-
ward and high-throughput
immu-
noaf
nity assays.
under the receiver operating characteristic (ROC)
curvemaynot be the only requirement for success-
ful use of biomarkers in clinics. Instead, based on
disease character and the cost of the follow-up
examination, biomarkers with either higher sensi-
tivity or higher speci
in blood plasma has the challenge of searching
through 10
13
aminoacids.
7
Use of altered post-
translational modi
cations and protein isoforms
as biomarkers would be an even more chal-
lenging undertaking due to the even higher
complexity and dynamic turnover of post-
translational modi
city may be preferable.
4
Different types of genetic sequence features or
biomolecules, such as gene mutations, SNP vari-
ants, mRNA transcripts, and metabolites can be
used as disease biomarkers. There is a clear
advantage to using proteins as biomarkers, and
this advantage stems from the diversity of
proteins. There is an estimated number of
20,300 genes,
1
7,900 unique metabolites,
5
approx-
imately 100,000 mRNA transcripts, and up to 1.8
million different protein species, if we consider
post-translational modi
cations (PTMs). For this
reason, the majority of protein biomarker studies
are still focused on the search for altered protein
concentrations in biological samples.
Identi
cation of proteins in cells, tissues, and
biological
fluids is dominated by mass spectro-
metry
e
based techniques, even though protein
and antibody arrays found their own niches.
8
e
10
At the protein identi
cation phase of a biomarker
development pipeline, several thousand
protein species are detected in a limited number
of biological samples. Relative quanti
cations.
6
Being the ulti-
mate products of gene expression, proteins
re
fication,
approaches are then used to compile a short
list of candidates for veri
ect multiple genomic and transcriptomic
alterations in their sequences, post-translational
modi
cation in the indepen-
dent set of clinical samples.
Biomarker veri
cations, and cellular abundance level.
A fraction of proteins is secreted into blood and
biological
cation is an important step to
exclude false positive discoveries made due to
the biological and technological bias introduced
at the identi
fluids and can thus be detected by
noninvasive diagnostic tests. The immense diver-
sity of protein species increases the chance to
identify a marker, or a panel of markers, for
each disease state. The diversity of protein vari-
ants, however, signi
cation phase. Assays used for veri-
fication, such as enzyme-linked immunosorbent
assays (ELISA) and selected reaction monitoring
(SRM),
11
provide accurate and reliable compar-
ison of protein levels in dozens to hundreds of
clinical samples.
Validation of protein biomarkers includes
testing their performance in very large cohorts
of clinical samples. Such studies employ stan-
dardized preclinical protein assays, rigorous
blinded analysis, and multicenter collaborative
cantly increases the analyt-
ical
challenge
of
correct
detection
and
measurement of a speci
c variant in biological
samples. For example, detection of a particular
nucleotide in the genome of a cell should meet
the analytical challenge of searching through
3.2
10
9
nucleotides, while the detection of
a speci
c amino acid in interleukin 6 sequence
Search WWH ::

Custom Search