Biology Reference
In-Depth Information
On-chip amplification and assembly
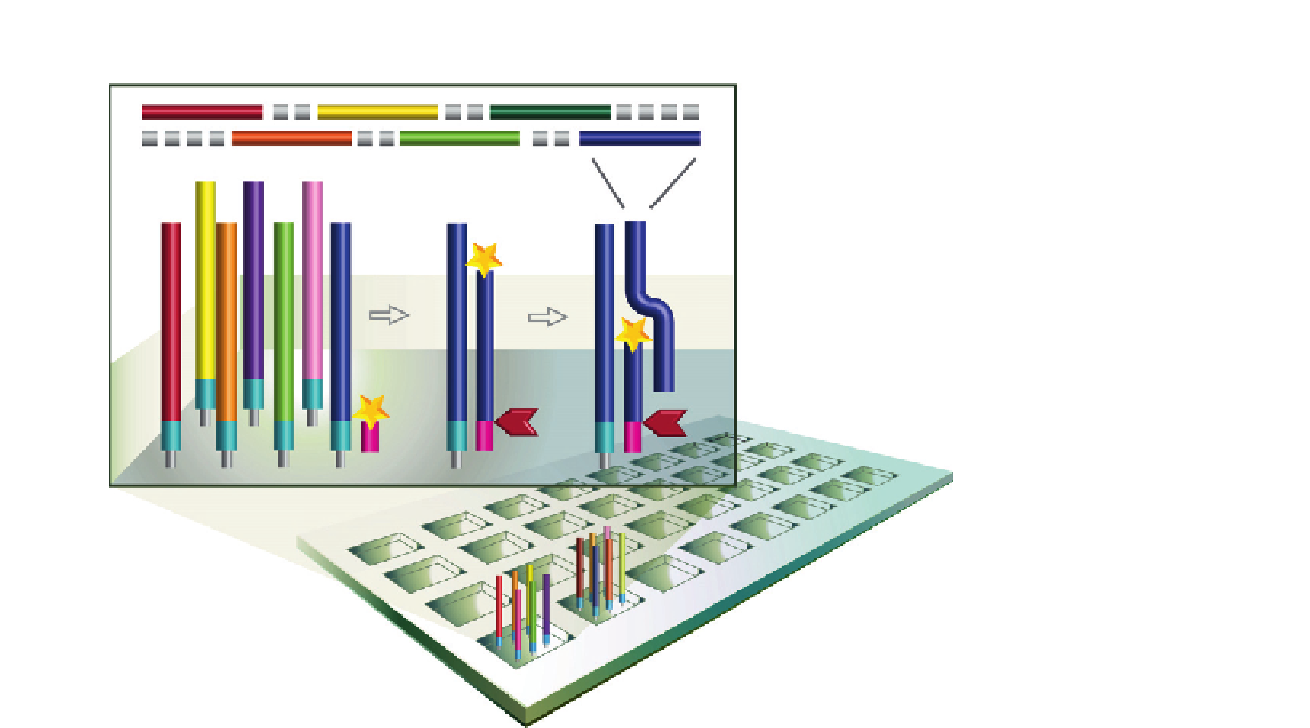
FIGURE 1.3
Integrated on-chip DNA
amplification and gene
assembly. The microchips
are divided into physically
isolated sub-arrays where
oligonucleotides are
amplified by isothermo
nicking and strand
displacement amplification.
The released strands are
the assembled into
0.5
Enbolished microchip
with endented subarrays
1 kb gene fragments
within the wells.
11
QUALITY CONTROL
Eliminating errors is critical for most gene synthesis projects, and is probably the most
costly and time-consuming step in gene synthesis. Errors are generated mainly during
chemical oligonucleotide synthesis and also in subsequent gene assembly steps.
In order to reduce the number of faulty oligonucleotides, chromatographic purification
by high-performance liquid chromatography (HPLC) or polyacrylamide gel electrophoresis
(PAGE) purification is typically used by conventional oligonucleotide synthesis vendors.
However, chromatographic purification is expensive and time-consuming, and PAGE
purification is even more costly. In this section, we will detail alternative purification
methods that couple with high-throughput synthesis technologies.
Fluorescence Selection
Reporter genes can be used to select in-frame gene fusions. Kim et al. used a fluorescence
selection method to improve the fidelity of gene synthesis by focusing on the elimination
of deletions and insertions in chemically synthesized genes. The authors constructed vectors
where GFP is expressed if the insertion of an in-frame DNA construct into appropriate
cloning sites shifts the GFP gene in frame.
64
The absence of insertions or deletions results
in fluorescent cell colonies when transformed into
E. coli
cells, which can be selected using
a transilluminator. The authors synthesized six similar-length genes from overlapping
oligonucleotides using ligase-based assembly and subsequent PCR, ligated the genes into
pBK reporter vectors, and transformed for overnight growth of colonies on agar plates.
On average, error rates improved from 1 in 629 bp (blind selection) to 1 in 6552 bp
(green fluorescence selection). This error rate allows for the straightforward construction
of error-free genes larger than 1000 bp by one-cycle synthesis.