Biology Reference
In-Depth Information
When grown independently, each population was unable to produce a significant amount
of fluorescent protein (either RFP or GFP). However, when both populations were grown in
separate chambers, which allowed passage of the QS signals but not cells, both GFP and
RFP were observed in the mixed population. That is, the passage of a QS signal between
populations was sufficient to trigger the expression of both fluorescent proteins. The authors
then sought to determine if both RFP and GFP could be expressed when both
E. coli
populations formed a biofilm. When either population was grown independently as a
biofilm, the biofilms were viable for up to two weeks. However, neither population
expressed its respective fluorescent protein. However, when both populations were grown
together in one biofilm, both GFP and RFP were detected within 24
36 hours of
inoculation. This behavior was stable for
6 days, whereupon the biofilm grew to exceed
its experimental limit (i.e. where oxygen diffusion began to affect fluorophore expression).
As such, this study demonstrates that a synthetic consortium can be engineered to exist as a
biofilm, and that the behavior of such biofilms is consistent over extended periods of time.
B
Recently, Brenner and Arnold created a synthetic consortium that could self-organize into a
structurally layered biofilm.
36
Their synthetic system consisted of two populations, each
labeled with a different fluorescent protein for ease of identification (
Fig. 13.3
). The first
population consisted of an
E. coli dapD
auxotroph (i.e. cannot synthesize the essential
metabolites diaminopimelate and lysine) labeled with a cyan fluorescent protein (CFP).
In this strain, the authors implemented a simple gene circuit that expresses
dapD
only in the
presence of the QS signal, C4HSL. When grown in the absence of C4HSL, this population
initially forms a healthy biofilm, but then quickly dies due to a lack of the aforementioned
metabolites. A second population of
E. coli
, labeled with a yellow fluorescent protein (YFP),
was engineered to lack the ability to form healthy biofilms by deleting genes required for
the expression of curli, type I pili, colanic acid, and capsular polysaccharides. Furthermore,
the authors engineered this strain to constitutively express C4HSL. When equal amounts of
these populations were grown in biofilm flow chambers, the two populations grew as a
highly intertwined biofilm. Here, the YFP population rescued the CFP population by
supplying C4HSL, thus activating
dapD
expression. Conversely, the CFP population allowed
the YFP population to form a biofilm, which consisted of an entangled biomass of both
YFP and CFP cells.
251
After 80 hours of growth, the authors observed that the biofilm would form a well-defined
structure where the YFP cells formed aggregates around CFP cells. As the biofilm matured,
the biofilm became stratified. Here, the CFP cells formed a uniform layer over the substrate,
while the YFP cells formed an uneven layer attached to the layer of CFP cells. Note that YFP
P
RHL
Biofilm
dapD
Δ
DapD
Biofilm
P
JM2300
C4HSL
Δ
Biofilm
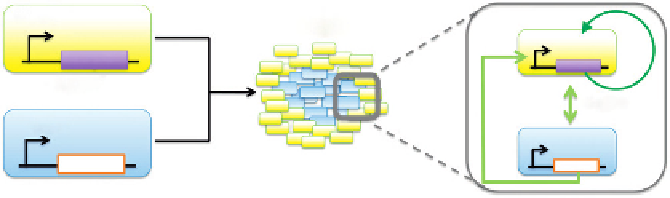
FIGURE 13.3
Engineering synthetic biofilms. To engineer a synthetic biofilm, Brenner and Arnold
36
created two strains that communicate
via a QS signal. The yellow population contains a C4HSL responsive promoter (P
RHL
), that when activated, drives expression
of dapD thus rescuing the population from death (left panel). The blue population constitutively expresses C4HSL via a
P
JM2300
promoter, but cannot make biofilms. When the populations are grown together, a biofilm forms where a population
of yellow cells surrounds a population of blue cells (center panel), which eventually leads to the formation of a structurally
layered biofilm (not shown). Within the biofilm, the C4HSL from the blue population rescues the yellow population, allowing
growth (green arrow) by driving expression of dapD while the yellow population allows the blue population to form a biofilm.