Biology Reference
In-Depth Information
(B)
Segments carrying no DNA
replication start sites
Segments carrying
DNA replication start sites
5'
5'
3'
Protein complex in
membrane, binds dsDNA,
introduces nick
3'
5'
3'
5'
5'
One ssDNA passes through
membrane into cytoplasm
where recA-mediated
homologous recombination
proceeds
3'
5'
Integrated in the
B. sustilis
genome
Established as plasmid
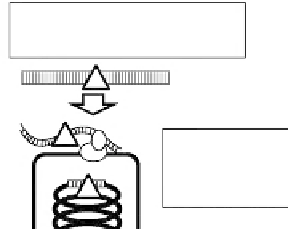
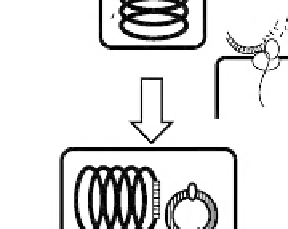
FIGURE 12.2B
B. subtilis transformation provides two DNA forms through natural competent cells. The inherent nature of B. subtilis 168 is
illustrated and briefly described in the text.
228
prokaryotic and eukaryotic hosts for genome synthesis applications will also be argued for
sporadically in the sections below.
THE KEIO METHOD
How Does B. subtilis Carry out DNA Incorporation and Connection?
Why was
B. subtilis
chosen as a host? The strain was known to be able to develop natural
competence,
13,14
an inherent remarkable feature, and conduct high and precise natural
homologous recombination.
1,12
As illustrated in
Figure 12.2B
,
B. subtilis
forms protein
complexes in membrane, responding to the quorum-sensing molecular mechanism, that are
able to trap double-stranded DNA (dsDNA) present outside. One of the proteins introduces
a nick in the bound exogenous dsDNA. Single-stranded DNA (ssDNA) starting at the nick is
taken up into the cytoplasm. The cleavage site by the nuclease and the strand selection by
the transformation complex are basically random.
13,14
The resultant highly recombinogenic
ssDNA is immediately recruited by cellular
recA
product and integrated into a
recA
-
dependent homologous recombination pathway.
14
The DNA promptly recombines with the
counter homologous sequences if present in the genome (
Figs 12.2A and 12.2B
). The
genetically coordinated molecular mechanism to transfer DNA actively through membranes
is absent in
E. coli
and yeast. Direct delivery of dsDNA to cytoplasm of both species has
to be catalyzed by physicochemical treatment, in sharp contrast to the inherent properties of
B. subtilis
.
15
A Genome Vector Suited for Giant DNA Handling
Use of
B. subtilis
168 as a host for stable guest cloning has a long history. BGM vectors are
B. subtilis
hosts specified to giant DNA cloning.
1,3,12
The name BGM, standing for
Ba
cillus
G
eno
M
e, was given to strains possessing accessory sequences in their genomes.
16
Accessory
sequences are used for selection in cloning processes illustrated in
Figure 12.2A
.