Biomedical Engineering Reference
In-Depth Information
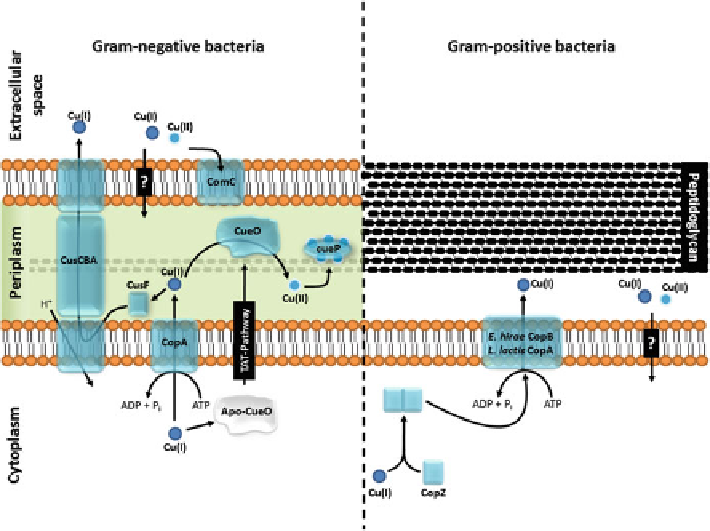
Fig. 6.1 Copper homeostasis and resistance mechanisms from Gram-negative and -positive
bacteria. In Gram-negative bacteria (from the
left
to the
right
): CusCBA extrudes Cu(I) from the
periplasm, CusA is a member of the resistance-nodulation-division (RND) protein superfamily of
proton-driven cation antiporters, CusC is an outer membrane factor (OMF), and CusB belongs to
the family of membrane fusion proteins (MFP), CusF is a copper chaperone that directs the copper
to the CusCBA efflux system [
30
]; CopA is a P
1B
-type ATPase that expels Cu(I) to the periplasm
with ATP hydrolysis [
75
]; ComC is an outer membrane protein that reduces copper permeability
by copper scaffolding; CueO is a multicopper which oxidizes periplasmic Cu(I) to Cu(II) [
34
];
CueP functions as a periplasmic-copper-pool. In Gram-positive (from the left to the right): CopZ is
a copper chaperone that binds to cytoplasmic copper and delivers it to the P
1B
-type ATPase
(
L. lactis
CopA/
E. hirae
CopB) [
69
], which in turn, extrudes Cu(I) outside of the cell [
80
]
chaperon CusF binds Cu(I) and delivers it to the CusCBA complex [
30
]. In
Salmonella enterica
sv.
Typhimurium
, a copper binding protein (CueP) (Fig.
6.1
),
under control of CueR, functions as a copper-pool, protecting the periplasm from
free-copper toxicity [
70
]. In a recent study it was shown that periplasmic and
cytoplasmic concentrations of copper in methylotrophic bacteria were higher in
the absence of ComC (copper-induced outer membrane component) protein,
highlighting its involvement in copper permeability (Fig.
6.1
). When this protein
was not present, copper concentrations were higher inside the periplasm and
cytoplasm, functioning as scaffolding or tethering protein in
E. coli
outer mem-
brane. The expression of this protein was shown to be controlled by ComR, a novel
TetR-like copper-responsive repressor [
55
].
Search WWH ::

Custom Search