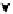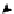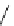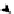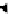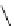Biology Reference
In-Depth Information
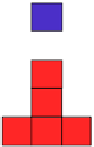
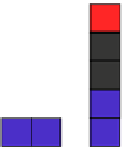
FIGURE 5.2
Inferring network structure from
transcriptional signatures. A, B, C: Given the struc-
ture and knowledge regarding the flow of information
through a network of n components that transduce the
input signal (IS) received by a cell, one can predict the
effects of perturbing individual network components on
the expression of known target genes (E). For example,
given the linear structure and flow of information of the
network shown in A, perturbing any of the network
components will affect the expression of target genes
E1
e
E6 in a similar manner as depicted in the sche-
matized heat map. D: Given a set of gene expression
signatures generated by perturbing the components of
a network, it should be possible to reconstruct the
structure of the network and deduce the flow of infor-
mation through its resident components.
A
B
IS
IS
N1
N1
N2
N2
E5
E6
N3
N3
E1
E2
E3
E4
E5
E6
E1
E2
E3
E4
N1
N2
N3
N1
N2 N3
E1
E1
E2
E3
E4
E5
E6
E2
E3
E4
E5
E6
C
D
IS
N1
N1
?
N2
N3
N2
N3
E1
E2
E5
E6
N4
N4
E3
E4
N1
N2 N3 N4
N1
N2 N3 N4
E1
E2
E3
E4
E5
E6
E1
E2
E3
E4
E5
E6
differentiation-correlated signature genes in leukemia cells.
This study identified eight bioactives that were further
validated as bona fide differentiation inducers.
The success of the first GE-HTS
[249]
led to the devel-
opment of the Luminex xMAP technology suitable for HTS
(see
Box 5.2
for details on the Luminex technology). The
technology combines multiplex ligation-mediated amplifi-
cation
[250
252]
with optically tagged and barcoded
microsphere and flow cytometric detection. The technology
is currently capable of measuring up to 500 transcripts
e