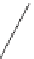Biology Reference
In-Depth Information
(A)
out-degree
in-degree
k
out
k
in
Hubs
k
degree distribution
(B)
(C)
(D)
flux-capacity
hierarchy
modularity
Modules
Hub
Bottleneck
GRN building blocks/motifs
(E)
Autoregulation
TF cascade
Single input motif
Bifan
Feed-forward loop
Feedback loop
Multi input motif
FIGURE 4.3
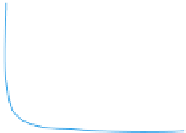
GRN structure and topological features. (A) The degree of a node refers to the number of connections it engages in. In GRNs, two types
of degree can be distinguished: the out-degree (k
out
) (left) refers to the number of genes targeted by a TF, and the in-degree (k
in
) (middle) reflects the
number of TFs that target a gene. All degrees in a GRN can be analyzed together to gain insight into the degree distribution (right). Nodes with a very high
degree are referred to as hubs. (B) The product of the in- and out-degrees is referred to as the flux-capacity, and illustrates the number of information paths
that pass through a node. (C) GRNs of E. coli and yeast are hierarchical, with different layers of TFs regulating one another. (D) GRNs are modular and
bottleneck TFs with high between-ness (the number of paths it engages in that connect any two nodes in the network). For visualization purposes, the
network is visualized with a single type of node (TF) and undirected edges (as in protein
e
protein interaction networks). (E) GRNs contain many different
types of small circuits, or building blocks. When statistically overrepresented, these circuits are referred to as network motifs.
but that some TFs are broadly involved in gene control.
Conversely, most genes are bound or regulated by a modest
number of TFs, while some exhibit extensive regulatory
input. Overall, the presence of hubs ensures network
connectivity and integrity
[125]
. By combining the in- and
the out-degree (k
in
xk
out
) the flux-capacity of a node can be
computed, which indicates the number of information paths
that pass directly through a node and a high flux-capacity
may indicate the importance of that node to network func-
tionality (
Figure 4.3
B)
[106]
. So far, the analysis of flux-
capacity has been performed only on an integrated network
that combines transcriptional regulation with post-tran-
scriptional regulation by microRNAs in C. elegans
[106]
.In
purely transcriptional GRNs (i.e., only involving TFs), flux-
capacity implies a hierarchical structure, with TFs as both
upstream regulators and downstream targets (
Figure 4.3
C).